
Identification of an optimal mutant allele frequency to detect activating KRAS, NRAS, and BRAF mutations in a commercial cell-free DNA next-generation sequencing assay in colorectal and pancreatic adenocarcinomas
Highlight box
Key findings
• Increasing mutant allele frequency (MAF) in cell-free DNA (cfDNA) sequencing predicts sensitivity for KRAS, NRAS, and BRAF mutations in colorectal and KRAS mutations in pancreatic adenocarcinoma.
What is known and what is new?
• Sequencing of cfDNA offers a convenient, safe, and relatively rapid method to assess a tumor’s molecular profile. However, sensitivity may be limited by low tumor DNA shedding into the plasma.
• This analysis uses MAF as a surrogate for tumor DNA content in a commercial cfDNA assay to predict sensitivity for clinically important mutations in colorectal and pancreatic ductal adenocarcinoma.
What is the implication, and what should change now?
• These data demonstrate practical and easily applied MAF thresholds to determine cfDNA assay adequacy to detect KRAS, NRAS, and BRAF mutations that may affect treatment selection. If an assay is deemed adequate, a patient may proceed to appropriately selected treatment more quickly or avoid an invasive tissue biopsy.
Introduction
In colorectal cancer (CRC), activating single-nucleotide variant (SNV) mutations in KRAS, NRAS, and BRAF are common (1-3), present in 39–44%, 4–8%, and 5–9% of patients, respectively. Therapy with anti-epidermal growth factor receptor (EGFR) monoclonal antibodies (mAb) alone or in combination with chemotherapy is an important therapeutic option in RAS/RAF wild-type (WT) CRC (4,5). Retrospective analyses comparing anti-EGFR therapy with the anti-vascular endothelial growth factor (VEGF) mAb bevacizumab in combination with chemotherapy have suggested superior efficacy for anti-EGFR therapy in left-sided metastatic RAS/RAF WT CRC as first line therapy (6), which has recently been confirmed in results from the prospective randomized PARADIGM trial (7). In contrast, the presence of activating SNVs in KRAS/NRAS and BRAFV600predict resistance to anti-EGFR mAb (8,9) and alternative targeted therapies for BRAFV600 (10-12), and KRASG12C (13-15), are now available for patients progressing on first-line therapy. These data and the increasing availability of targeted treatments solidifies the importance of accurate ascertainment of RAS/RAF status prior to first line therapy in CRC, supported in national guidelines (16).
In pancreatic ductal adenocarcinoma (PDAC), 85–90% of patients harbor KRAS activating mutations (17), with a small fraction harboring targetable KRASG12C mutations (13,18,19). Patients with KRAS WT PDAC often have actionable fusion alterations (20-24), providing additional therapeutic options and spurring new treatment strategies. In addition, there are several novel molecular and immune-based approaches targeting KRAS-mutated cancers in development that will be of particular interest in pancreatic cancer given limited therapeutic options in this malignancy (25,26). Finally, KRAS mutational status carries prognostic significance in PDAC (27). Thus, accurate molecular testing to determine KRAS mutational status (although not NRAS or BRAF, as in CRC) is increasingly important in PDAC, both to allow access to novel targeted therapies and also because a negative KRAS result should prompt an extensive evaluation for alternative driver events, including rare fusions.
Sequencing of cell-free DNA (cfDNA) derived from plasma provides a convenient method to assay a tumor’s molecular profile (28). In comparison to tissue biopsies, advantages of sequencing cfDNA include avoidance of invasive procedures, speed to rapidly assess a tumor when urgent treatment is required, ease of sequential repeat assays over time, and assessment of the full spectrum of a cancer’s molecular profile. However, the sensitivity of cfDNA assays for actionable alterations is limited by the proportion of circulating tumor DNA (ctDNA) in the plasma (29). ctDNA content and thus sensitivity is affected by many factors, including volume of disease, location of metastases, and tumor specific factors (30). Overall, metastatic CRC exhibits high rates of detectable ctDNA in clinical practice, while PDAC has among the lowest rates of detectable ctDNA (31). However, CRC patients with metastases to either only lung or to the peritoneum have lower ctDNA content compared with liver metastases or more extensive multi-focal disease (32). Relatively low ctDNA shedding in PDAC is thought to be related to several factors, including low neoplastic cellularity and a highly desmoplastic stromal environment that restricts shedding (33).
In addition, cfDNA sequencing may detect mutations from sources other than the target malignancy. The detection of mutations in peripheral blood leukocytes, i.e., clonal hematopoiesis (CH), is a now well-documented phenomenon (34). Because the majority of cfDNA is derived from leukocytes, even very small CH clones may be detected on cfDNA next-generation sequencing (NGS) testing (35). While the most common CH mutations, such as DNMT3A and TET2, are specific to CH and thus easily recognized as myeloid in origin, mutations in TP53 and more rarely KRAS and BRAF, among others, are also seen in CH and may be difficult to distinguish from solid tumor-derived variants (35-39). The large majority of these confounding mutations are seen at a mutant allele frequency (MAF) <1% (35). Both low ctDNA shedding and potential confounding mutations present a challenge to assessing cfDNA assay adequacy.
To address this issue, Meador et al. examined sensitivity in cfDNA for EGFR and KRAS mutations in non-small cell lung cancer (NSCLC) based on the maximum allele frequency of other dominant variants in the cfDNA (40). Their data showed that cfDNA with dominant gene maximum allele frequency of >1% predicted 100% sensitivity for EGFR and KRAS mutations. Greater resolution was limited by small patient numbers (n=29) with maximum allele frequency below 1% and the assay itself was institution-specific, limiting broad applicability. Nonetheless, the idea of a simple quantitative threshold to predict assay sensitivity is appealing for the practicing oncologist. Similarly, Nakamura et al. proposed an identical threshold of >1% as sufficient to assess microsatellite instability in Guardant cfDNA assays (41).
Defining both mutated and WT status for RAS/RAF mutations is important for appropriate care in CRC and PDAC, however the sensitivity for oncogenic RAS/RAF SNVs in the cfDNA of these cancers based on non-RAS/RAF oncogenic variant MAF is not known. Thus, we explored a clinically applicable MAF threshold in the Guardant360 cfDNA commercial assay for detection of KRAS, NRAS, or BRAF oncogenic mutations in CRC and PDAC tumors. We present this article in accordance with the MDAR reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-114/rc).
Methods
Patient populations
Patient molecular and clinical data were obtained from an internal database of patients with CRC and PDAC undergoing treatment at Duke Cancer Institute (DCI) who received cfDNA NGS testing and from the SCRUM-Japan GOZILA study (42). Only patients with metastatic disease at the time of cfDNA testing were included. GOZILA is a plasma genomic profiling study involving 31 core cancer institutions in Japan. Patients with metastatic gastrointestinal cancers were eligible for GOZILA; patients with CRC and PDAC were selected for this study. Eligible patients for GOZILA provided written informed consent, including publication of any materials. To avoid the suppression of ctDNA shedding because of chemotherapy, the GOZILA patients were required to have disease progression following systemic chemotherapy and to have not yet started subsequent therapy at the time of blood sampling. For GOZILA, patients were enrolled and plasma genotyping took place between January 2018 and February 2021.
All comprehensive genomic profiling (CGP) tests performed at DCI were ordered at the discretion of oncology providers as a component of routine cancer care and were stored in the electronic medical record as well as an internally developed data warehouse solution called the Duke Molecular Registry of Tumors (MRT) (43). Approval for the establishment of MRT as a clinical and research data repository was granted by the IRB of the Duke University Medical Center (DUMC) (No. Pro00085260). Specific approval for this research collaboration and waiver of consent was also granted by the DUMC IRB (No. Pro00109863).
All included patients underwent tissue CGP with commercially available assays. Duke patients underwent tissue testing with FoundationOne CDx (Foundation Medicine). The GOZILA cohort underwent tissue testing with the Oncomine Comprehensive Assay (ThermoFisher). These tissue-based assays are multi-gene panels that cover and report genes of interest to this study, specifically activating mutations in KRAS and NRAS exons 2, 3, and 4, and BRAF. Guardant360 (Guardant Health) was the sole cfDNA assay used for both Duke and GOZILA patients.
Patients with CRC or PDAC with both tissue and cfDNA assays available were identified in the GOZILA and MRT databases, and tumors with oncogenic activating SNVs in KRAS, NRAS, or BRAF identified on tissue-based sequencing were selected for further analysis. Activating mutations in KRAS and NRAS exons 2-4 (codons 12, 13, 34, 61, 117, and 146), BRAF codon V600, and oncogenic, non-BRAF V600 Class II and Class III variants were included (see Table S1 for all included variants). Although BRAF Class II and Class III variants do not currently guide clinical decision making for anti-EGFR therapy, these variants have been shown to be biologically relevant in CRC and could potentially guide clinical trial enrollment, and were thus included in this study (44,45). In two instances in which an NRAS or BRAF non-V600 mutation was found with a KRAS mutation on the tissue sample, only the KRAS mutation was included in the cfDNA sensitivity analysis in order to avoid redundancy.
These studies were conducted in accordance with the Declaration of Helsinki (as revised in 2013) and the Japanese Ethical Guidelines for Medical and Health Research Involving Human Subjects. All study protocols were approved by the institutional review board of each participating institution and the GOZILA study was registered at the University Hospital Medical Information Network Clinical Trials Registry (No. UMIN000042612).
cfDNA assay characteristics
Guardant360 CDx is a cfDNA NGS assay that reports SNVs and insertions/deletions (Indels) for 74 genes, in addition to 18 copy number alterations and 6 fusion products not relevant to the current study (46). Guardant reports analytical sensitivity for SNVs and Indels of 99.7–100% based on MAF of ³0.5%, and 70.6–86% based on MAF <0.5% with 30 ng of input DNA. Guardant360 CDx limit of detection (LoD) is reported at 0.8–3% MAF per copy number at 5 ng input DNA and 0.1–0.8% MAF per copy number for 30 ng of input DNA.
Variant analysis
All variant analysis was performed by Guardant Health. Variants of uncertain significance (VUS) were excluded from the analysis. All clinically reported, non-VUS alterations were considered oncogenic and included in the analysis. Comprehensive germline variant testing was not available and so germline variants could not be definitively excluded. In addition, mutations in JAK2 were excluded as this gene is strongly associated with CH but lacks a known biologic role in CRC or PDAC. Other similar genes commonly associated with CH, such as DNMT3A and TET2, are not reported in the Guardant360 CDx assay (46).
Analytic plan
Each available cfDNA assay was treated independently. On each cfDNA assay, detection of the tissue RAS/RAF mutation was noted, and the non-RAS/RAF oncogenic mutation (“dominant” mutation) with the greatest MAF was identified. Assays with no detected non-RAS/RAF oncogenic alterations were considered MAF =0. The assays were then analyzed for sensitivity based on MAF. We also selected cfDNA NGS assays that did not successfully detect RAS/RAF mutations for further analysis. Of these, we identified assays with oncogenic variants present, and compared these variants to results from tissue NGS assays. Concordant and non-concordant variants were noted for each individual patient.
Statistical analysis
We employed recursive partitioning and a maximally selected rank statistic to identify optimal cutpoints of MAF for sensitivity. We then used the resulting optimal cutpoint to select ranges of dominant oncogenic mutation greatest MAF below 1% to calculate sensitivity for RAS/RAF SNVs, and predefined intervals of 1–5%, 5–10%, and >10% MAF for ranges above 1%. Recursive partitioning (https://cran.r-project.org/web/packages/rpart/index.html) splits the data by the MAF values and the proportion of the group where RAS/RAF SNVs are detected (47). Each cutpoint maximizes the number of detected RAS/RAF SNVs in one group vs. the group below the cutpoint. The maxstat package (https://cran.r-project.org/web/packages/maxstat/index.html) tests every possible cutpoint where reasonable sample sizes exist in each group and identifies the cutpoint at which the probability of RAS/RAF SNV detection is maximized using a Wilcoxon rank statistic (48). All analyses were done using R version 4.1.3 (49) (2022-03-10) within RStudio 2022.7.1.554 (50).
Results
Patient characteristics
Of 1,604 CRC and 572 PDAC patients enrolled on the GOZILA study, and 885 CRC and 442 PDAC patients from the Duke MRT, 165 total patients with 198 cfDNA assays met criteria and were included in the analysis (Figure 1). Clinical characteristics of this cohort, overall and by cancer type, as well as primary tumor site within the colon or pancreas, are shown in Tables 1,2. The GOZILA study contributed 108 of 135 CRC and the Duke MRT contributed 25 of 30 PDAC cases. Patients in the GOZILA cohort were more likely to have undergone resection of the primary tumor. Number and distribution of metastatic sites at time of initial cfDNA NGS testing is shown in Table S2.
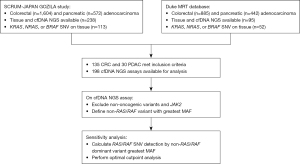
Table 1
| Characteristic | CRC + PDAC | CRC | PDAC |
|---|---|---|---|
| Full cohort, n | 165 | 135 | 30 |
| Duke MRT | 53 | 27 | 25 |
| GOZILA-SCRUM | 113 | 108 | 5 |
| Age at 1st treatment, median [range] | 60 [30–83] | 60 [30–83] | 62 [36–82] |
| Female, n (%) | 86 (52.1) | 72 (53.3) | 14 (46.7) |
| Race/ethnicity, n (%) | |||
| Asian | 115 (69.7) | 109 (80.7) | 6 (20.0) |
| Caucasian | 38 (23.0) | 17 (12.6) | 21 (70.0) |
| African-American | 11 (6.7) | 8 (5.9) | 3 (10.0) |
| Native American | 1 (0.6) | 1 (0.7) | 0 |
| Hispanic ethnicity | 0 | 0 | 1 (3.3) |
CRC, colorectal cancer; PDAC, pancreatic ductal adenocarcinoma; MRT, Molecular Registry of Tumors.
Table 2
| Primary site | Patients, n (%) |
|---|---|
| Colorectal primary | |
| Cecum | 18 (13.3) |
| Ascending | 25 (18.5) |
| Transverse | 11 (8.1) |
| Descending/sigmoid | 42 (31.1) |
| Rectal | 36 (26.7) |
| Unknown | 3 (2.2) |
| Primary resected, full cohort | 96 (71.1) |
| Duke MRT | 14 (51.9) |
| GOZILA-SCRUM | 82 (75.9) |
| Pancreatic primary | |
| Head | 15 (50.0) |
| Body | 7 (23.3) |
| Tail | 8 (26.7) |
| Primary resected, full cohort | 4 (13.3) |
| Duke MRT | 1 (4.0) |
| GOZILA-SCRUM | 3 (60.0) |
MRT, Molecular Registry of Tumors.
Twenty-two point two percent of patients with CRC received anti-EGFR mAb therapy at some point in their clinical course (Table S3), however all patients had comprehensive testing on tissue obtained prior to anti-EGFR therapy. Most of these patients had BRAF mutations and received anti-EGFR therapy either as part of a clinical trial, as SOC in combination with BRAF/MEK inhibitors, or harbored a BRAF Class II or III mutation that was not considered a contraindication to anti-EGFR therapy. Five patients received anti-EGFR despite predicted resistance, though the clinical reasoning was not readily discernable; in at least one instance CGP on a past tissue sample revealed a previously undetected KRAS mutation.
Patient molecular testing
Among CRC patients, KRAS SNV mutations were most common (98/135, 72.6%) followed by BRAF (35/135, 25.9%) and NRAS (4/135, 3.0%) (Table 3, Table S1). For the 2 patients with a concurrent KRAS and either BRAF or NRAS mutation, only the KRAS mutation was considered for the purposes of the primary analysis. All 30 patients with PDAC harbored KRAS mutations.
Table 3
| Characteristic | CRC + PDAC (n=165) | CRC (n=135) | PDAC (n=30) |
|---|---|---|---|
| Total cfDNA assays, n | 198 | 167 | 31 |
| Patients with 2+ assays, n (%) | |||
| 2 | 8 (4.8) | 7 (5.2) | 1 (3.3) |
| 3 | 11 (6.7) | 11 (8.1) | 0 |
| 4 | 1 (0.6) | 1 (0.7) | 0 |
| Time from tissue NGS to cfDNA NGS specimen collection | |||
| Complete data, n (%) | 176/198 (88.9) | 150/167 (89.8) | 26/31 (83.9) |
| Median days (range) | 293.5 (−712 to 2,512) | 315 (−316 to 2,512) | 10.5 (−712 to 1,403) |
| n within 90 days (%) | 34/176 (19.3) | 23/150 (15.3) | 11/26 (42.3) |
| Systemic therapy between tissue and cfDNA NGS, n (%) | |||
| Complete data | 172/198 (86.9) | 146/167 (87.4) | 26/31 (83.9) |
| Treatment | 143/172 (83.1) | 126/146 (86.3) | 17/26 (65.4) |
| RAS/RAF SNV on tissue, n (%) | |||
| KRAS | 128 (77.6) | 98 (72.6) | 30 (100.0) |
| NRAS | 4 (2.4) | 4 (3.0) | |
| BRAF | 35 (21.2) | 35 (25.9) | |
| RAS/RAF SNV present on cfDNA NGS, n (%) | 155/198 (78.3) | 138/167 (82.6) | 17/31 (54.8) |
| No oncogenic variants detected in cfDNA, n (%) | 25/198 (12.6) | 15/167 (9.0) | 10/31 (32.3) |
| Dominant mutation with greatest MAF, gene (% of detected) | TP53 (53.9) | TP53 (80.0) | |
| APC (21.1) |
cfDNA, cell-free DNA; NGS, next-generation sequencing; CRC, colorectal cancer; PDAC, pancreatic ductal adenocarcinoma; SNV, single nucleotide variant; MAF, mutant allele frequency.
One hundred and ninety-eight total cfDNA assays were included in the analysis. Twenty patients contributed 2 or more assays to the analysis (Table 3). Median time from reference tissue NGS to cfDNA NGS was 293.5 (range, −712 to 2,512) days for all assays, 315 (range, −316 to 2,512) days for CRC assays, and 10.5 (range, −712 to 1,403) days for PDAC assays. CRC, PDAC, and all assays combined had tissue and cfDNA NGS obtained within +/− 90 days in 15.3%, 42.3%, and 19.3% of assays respectively. The large majority of patients received systemic therapy between tissue and cfDNA assays.
No oncogenic, non-VUS alterations were detected in 9.0% of CRC and 32.3% of PDAC cfDNA assays (Table 3). TP53 variants were the most frequently detected mutations with greatest MAF. The RAS/RAF SNV detected on tissue testing was also detected in 82.6% of CRC cfDNA assays and 54.8% of PDAC cfDNA assays (Table 3). Number of metastatic sites and solitary site of metastasis stratified by successful detection of RAS/RAF SNV in cfDNA is shown in Table S2. Twenty point seven percent of CRC had only 1 site of metastatic disease at time of the cfDNA assay, and of these only 50% had successful detection of RAS/RAF. For CRC with 2 or sites of disease, detection of RAS/RAF SNVs was numerically greater at 86.8%. Forty-three point three percent of PDAC had 1 site of metastatic disease, with 61.5% successful detection of the KRAS SNV.
Sensitivity for RAS/RAF mutations by MAF
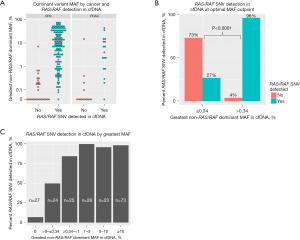
Oncogenic, non-RAS/RAF dominant oncogenic variants with the greatest cfDNA MAF were identified and patients were stratified by detection of the RAS/RAF SNV (Figure 1). Pairs of RAS/RAF and dominant mutations with associated MAF are shown in Table S1. The distribution of dominant variants with greatest MAF stratified by cancer and detection of RAS/RAF SNVs is shown in Figure 2A. Recursive partitioning and maximally selected rank statistic methods were employed to identify the optimal MAF discrimination for sensitivity for RAS/RAF SNVs while excluding the fewest assays, with both methods yielding an MAF of 0.34%. Cutpoints above 0.34% improve sensitivity marginally but exclude a greater portion of total assays. For example, 76 of 198 total assays (38.4%) fell below the MAF threshold at an MAF of 1% compared with 50 of 198 assays (25.3%) at MAF 0.34% (Figure S1). In contrast, cfDNA assays in which the RAS/RAF SNV was not detected rise sharply with dominant gene MAF below 0.34%, but exclude significantly fewer assays (Figure S1). Ninety-six percent of RAS/RAF SNVs were detected with MAF >0.34%, while only 27% were detected £0.34% (significant at P<0.0001 by Wilcoxon rank statistic, Figure 2B). Sensitivity analysis by ranges of greatest dominant MAF (Figure 2C) confirmed increasing sensitivity with increased dominant MAF; RAS/RAF sensitivity was >98% with MAF greater than 1%, fell to 84.0% for MAF between 0.34 and 1%, and was only 50.0% for MAF >0 and £0.34%.
Analysis of false negative cfDNA assays
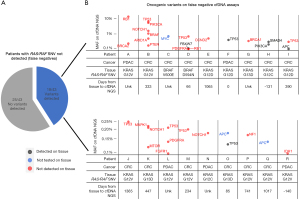
Next, the 43 cfDNA NGS assays in which the RAS/RAF SNVs were not detected (i.e., false negatives) were examined. Oncogenic variants were detected in 18 of these 43 cfDNA assays while the remaining 25 had no oncogenic variants detected (Figure 3A). For the 18 patients with detected oncogenic variants, the individual assay with the detected cfDNA variants, associated cancer, tissue KRAS, NRAS, or BRAF SNV, and time between tissue and cfDNA NGS are shown in Figure 3B. From these 18 cfDNA assays, 25 of 34 total detected variants and 13 of the 18 variants with the greatest MAF were not detected on tissue NGS (Figure 3B). Only 6 of 34 detected variants were also found on tissue NGS, while 3 variants (1 MYC and 2 APC mutations) seen on cfDNA were not tested on the tissue assay (Figure 3B). Only 5 mutations had MAF >1%, 4 from one patient, none of which were detected on tissue. TP53 was the most frequently mutated gene not also detected on tissue, however >1 mutation in RB1, NOTCH1, and PDGFRA were also observed.
Discussion
Despite the several advantages of cfDNA NGS, assay sensitivity represents a practical challenge for a clinical oncologist. In particular, defining mutational status of a gene, such as KRAS, is important for clinical decision making, however the absence of a mutation raises the question of whether the mutation was present but not detected. In this study, we define thresholds of greatest MAF of dominant non-RAS/RAF mutations for sensitivity for RAS/RAF SNVs in CRC and KRAS SNVs in PDAC. Based on these findings, we propose a practical approach to predicting the adequacy of the Guardant360 CDx cfDNA assay in CRC and PDAC (Figure 4). If a RAS/RAF SNV is detected, the assay may be considered accurate. If no oncogenic variants are detected, the assay is inadequate and alternative or repeat testing may be needed. If oncogenic variants are detected, but no RAS/RAF mutation, the oncogenic variant with the greatest MAF may predict if the assay is sufficiently sensitive. Specifically, dominant gene MAF >1% suggests very high sensitivity, while MAF >0.34% and <1% also retains moderately high sensitivity. Below MAF of 0.34%, the assay should be considered insufficiently sensitive to determine WT status.
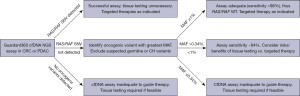
These thresholds may grant increased confidence around the sensitivity of the Guardant360 CDx assay and may have significant implications for anti-EGFR therapy or rechallenge in CRC, targeted therapies against KRASG12C or BRAFV600E, and the necessity of additional biopsy. In PDAC, high confidence that a patient is truly KRAS WT may prompt more extensive testing, especially to evaluate for rare but often actionable fusions. In addition, this analysis may inform use of cfDNA NGS assays to define inclusion and exclusion criteria for clinical trials. Relying solely on cfDNA may provide the most rapid assessment of a tumor to minimize time to treatment and avoids the need for invasive biopsy. However, in the case of a negative or inadequate cfDNA assay, the utility of repeat cfDNA NGS is unknown and best answered in the context of a trial.
An important limitation of our study is the use of a sole commercial cfDNA assay. Other assays and vendors have different testing characteristics and the technology itself may evolve over time, limiting lessons drawn specifically from this study. An advantage of this commercial assay is broad availability and use in the community; in contrast some prior studies (40) have used institutional assays or technologies associated with clinical trials that the clinicians may not access. In addition, while the technical specifications of cfDNA NGS assays may vary, we believe we have demonstrated important inherent characteristics of these assays.
Interestingly, among cfDNA assays that failed to detect the RAS/RAF SNV, we found that 25 of 31 (80.6%) of oncogenic variants were unique to the cfDNA assay and not detected on tissue testing. In addition, the MAF was <1% for nearly all of these variants. We speculate that this observation may be due to detection of variants arising from intra-tumoral heterogeneity, clonal evolution of the tumor, secondary malignancy, or CH. Of these, the contribution of CH is the most intriguing and problematic for interpretation of cfDNA assays. Leukocytes are thought to contribute the largest fraction of plasma cfDNA and detection of even low-level clonal variants in leukocytes is well-described and unsurprising (35). The most common variants associated with CH such as DNMT3A, TET2, and JAK2 are easily recognized and excluded (34). However, mutations in many genes important in solid malignancies such as TP53 are also regularly observed in leukocytes (36,37) and may confound accurate assessment of a tumor.
Unfortunately, our study did not have access to leukocyte sequencing to confirm the degree of confounding from CH. We suspect that sensitivity assessment and thus assay reliability may be improved with leukocyte variant subtraction but would add greater cost and greater burden of interpretation. Whether a strategy of leukocyte sequencing and subtraction is cost-effective or clinically meaningful requires further study. This finding further suggests that these assays may not be limited by poor sensitivity inherent to the assay itself at MAF <1%, but rather confounding from low level variants. Instead, many of the patients in our study with greatest MAF <1% may actually have levels of ctDNA below the limits of the assay. An important implication of this confounding is that improving the technical limits of a cfDNA assay, whether the Guardant360 assay used in this study or an alternative, for example by increasing read depth or DNA input, is also more likely to identify very small clonal variants. These low MAF variants may then be difficult to interpret for clinical actionability, given the increased likelihood these are leukocyte-derived. Our data does show that sensitivity for clinically relevant tumor-derived mutations improves with increasing greatest dominant gene MAF and provides a practical rule for assessing assay adequacy despite the potential presence of confounding mutations.
Conclusions
In this study, we show that greatest MAF of an oncogenic variant is a useful proxy for ctDNA content and to assess that an assay is sufficiently sensitive to detect clinically important mutations in KRAS, NRAS, and BRAF. In cases where tissue-based molecular testing is unavailable or difficult to obtain, cfDNA assay with adequate ctDNA may allow more rapid clinical decision making and help guide or avoid the need for invasive biopsy. In addition, low concordance between variants detected on false negative assays and tissue assays in our study suggest increased risk of confounding or heterogeneity when ctDNA content is low, though further research is needed to adequately explore this finding. These data, building on prior studies, increase the practicing oncologist’s confidence to interpret and effectively use cfDNA NGS assays in clinical care.
Acknowledgments
A version of this work was previously presented at the American Society of Clinical Oncology Annual Meeting in Chicago, IL, on June 4, 2022.
Funding: This work was supported by SCRUM-Japan, who provided financial support for the GOZILA and GI-SCREEN studies and for data aggregation; and the Duke Cancer Institute who provided internal funds for data storage and aggregation.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-114/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-114/dss
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-114/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-114/coif). KU reports honoraria from Chugai Pharmaceutical, Taiho Pharmaceutical and Yakult Honsha. MG reports royalties from SciMed Solutions as part of a software licensing agreement. MI reports grants/research funding from Eisai, Merck biopharma, Eli Lilly Japan, Yakult, Ono, ASLAN, J-Pharma, AstraZeneca, Pfizer, Merus N.V., Nihon Servier, Delta-Fly Pharma, Chiome Bioscience, Bristol-Myers Squibb, Chugai Pharmaceutical, Novartis, Bayer, Takeda, MSD, Syneos Health, as well as honoraria from Eisai, MSD, Eli Lilly Japan, Yakult, Teijin Pharma, Astellas, Sumitomo Dainippon, Otsuka, Nihon Servier, Taiho, Chugai Pharmaceutical, Bristol-Myers Squibb, Novartis, Bayer, Takeda, EA Pharma, AstraZeneca, AbbVie, Abbott Japan, Fujifilm Toyama Chemical, and Incyte Biosciences Japan. MU reports grants/research fundings from Taiho Pharmaceutical, AstraZeneca, Merck, MSD, Astellas Pharma, Eisai, Ono Pharmaceutical, Incyte, Chugai Pharmaceutical, DFP, Daiichi Sankyo, Novartis, Boehringer Ingelheim, and J-pharma, and also honoraria from Taiho Pharmaceutical, AstraZeneca, Yakult Honsha, MSD, Nihon Servier, Ono Pharmaceutical, Uncyte, Chugai Pharmaceutical, Boehringer Ingelheim, and J-Pharma. HT reports grants/research funding from Daiichi Sankyo and Takeda, and also honoraria from Guardant Health AMEA, Chugai Pharmaceutical, Ono, Eli Lilly, Takeda, and Merck. YK reports scholarly/research support or grant funding from Ono, Chugai Pharmaceutical, Taiho, Shionogi, Nippon Zoki, Asahi Kasei, Nippon Kayaku, Daiichi Sankyo, IQVIA Services Japan, MSD, Astellas Pharma, Incyte, Eisai, National Cancer Center Japan, Syneos Health, ShiftZero, Parexel International, Japan Clinical Cancer Research Organization, EPS, Sysemx Corporation, and the Public Health Research Foundation; and also honoraria from Ono, Taiho, Astellas Pharma, EA Pharma, Daiichi Sankyo, Nippon Kayaku, Pfizer, Nippon Zoki, Sanofi, Nipro, Asahi, Chugai Pharmaceutical, MSD, Zeria, Eli Lilly, Bayer, Yakult, Sumitomo Dainippon Pharma, Incyte, and Merck. TE reports research grants and honoraria from MSD, Daiichi Sankyo, and, Chugai, grants from Pfizer, Astellas, Quintiles, Syneos Health, Amgen, Ono, Novartis, Astellas, Amgen Biopharma, Asahi Kasei Pharma, and IQVIA, and honoraria from Taiho, Eli Lilly, and Bristol Myers Squib, outside the submitted work. TD reports research funds from MSD, Amgen and Ono Pharmaceutical, and honoraria from Sysmex, Ono Pharmaceutical, SAWAI Pharmaceutical Co and Daiichi Sankyo, outside the submitted work. HB reports research funding from Ono Pharmaceutical and honoraria from Ono Pharmaceutical, Taiho, and Eli Lilly. TBS reports research funding/grants from Agio, Arys, Arcua, Atreca, Boston Biomedical, Bayer, Eisai, Celgene, Lilly, Ipsen, Clovis, Seattle Genetics, Genentech, Novartic, Mirati, Merus, Abgenomics, Incyte, Pfizer, and BMS; consulting fees from Ipsen, Arcus, Pfizer, Seattle Genetics, Bayer, Genentech, Incyte, Eisai, Merus, Merck, Stemline, Abbvie, Boehringer Ingelheim, Janssen, Daiichi Sankyo, Natera, TreosBio, Celularity, Caladrius Biosciences, Exact Science, Sobi, Beigene, Kapaph, AstraZeneca, Deciphera, Zai Labs, Exelixis, MJH Life Sciences, Aptitude Health, Illumina, Foundation Medicine, and Sanofi; patents currently licensed to Imugene and Recursionl; and participation in DSMB or Scientific Advistory Boards for The Valley Hospital, Fibrogen, Suzhou Kintor, AstraZeneca, Exelixis, Merck, Eisai, PanCan, 1GLOBE, Imugene, Immuneering, Xilis, Replimune, Artiva, and Sun Biopharma. TY reports research support/grants from Amgen, Chugai Pharmaceutical, Daiichi Sankyo, Eisai, FALCO, Genomedia, Molecular Health GmbH, MSD, Boehringer Ingelheim, Ono, Pfizer, Roche Diagnostics, Sanofi, Sysmex Corporation, and Taiho as well as consulting fees from Sumitomo Corporation and honoraria from Bayer, Chugai Pharmaceutical, Merck, MSD, Ono, and Takeda. JS reports grant/research support from Abbvie, Amgen, AStar D3, Bayer, Beigene, Curegenix, Daiichi-Sankyo, Eli Lilly, Erasca, Gossamer Bio, Leap Therapeutics, Nektar, Roche/Genentech, Seagen, and Silverback Therapeutics; and consulting fees from or serving in an advisory role to Abbvie, Amgen, AstraZeneca, Bayer, Beigene, Daiichi-Sankyo, Eli Lilly, G3 Therapeutics, GSK, Natera, Pfizer, Pionyr Immunotherapeutics, Roche/Genentech, Seagen, Silverback Therapeutics, Takeda, and Viatris. YN reports grants and honoraria from Chugai, grants from Taiho, Guardant Health, Genomedia, Daiichi-Sankyo, Seagen, and Roche Diagnostics, and honoraria from Guardant Health AMEA and Merck. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and the Japanese Ethical Guidelines for Medical and Health Research Involving Human Subjects. All study protocols were approved by the institutional review board of each participating institution and the GOZILA study was registered at the University Hospital Medical Information Network Clinical Trials Registry (No. UMIN000042612). Approval for the establishment of the Duke MRT as a clinical and research data repository was granted by the Duke University Medical Center (DUMC) IRB (No. Pro00085260). Specific approval for this research collaboration and waiver of consent was also granted by the DUMC IRB (No. Pro00109863).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330-7. [Crossref] [PubMed]
- Yaeger R, Chatila WK, Lipsyc MD, et al. Clinical Sequencing Defines the Genomic Landscape of Metastatic Colorectal Cancer. Cancer Cell 2018;33:125-136.e3. [Crossref] [PubMed]
- Zaidi SH, Harrison TA, Phipps AI, et al. Landscape of somatic single nucleotide variants and indels in colorectal cancer and impact on survival. Nat Commun 2020;11:3644. [Crossref] [PubMed]
- Venook AP, Niedzwiecki D, Lenz HJ, et al. Effect of First-Line Chemotherapy Combined With Cetuximab or Bevacizumab on Overall Survival in Patients With KRAS Wild-Type Advanced or Metastatic Colorectal Cancer: A Randomized Clinical Trial. JAMA 2017;317:2392-401. [Crossref] [PubMed]
- Douillard JY, Oliner KS, Siena S, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med 2013;369:1023-34. [Crossref] [PubMed]
- Arnold D, Lueza B, Douillard JY, et al. Prognostic and predictive value of primary tumour side in patients with RAS wild-type metastatic colorectal cancer treated with chemotherapy and EGFR directed antibodies in six randomized trials. Ann Oncol 2017;28:1713-29. [Crossref] [PubMed]
- Watanabe J, Muro K, Shitara K, et al. Panitumumab vs Bevacizumab Added to Standard First-line Chemotherapy and Overall Survival Among Patients With RAS Wild-type, Left-Sided Metastatic Colorectal Cancer: A Randomized Clinical Trial. JAMA 2023;329:1271-82. [Crossref] [PubMed]
- Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 2008;26:1626-34. [Crossref] [PubMed]
- Di Nicolantonio F, Martini M, Molinari F, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol 2008;26:5705-12. [Crossref] [PubMed]
- Kopetz S, Grothey A, Yaeger R, et al. Encorafenib, Binimetinib, and Cetuximab in BRAF V600E-Mutated Colorectal Cancer. N Engl J Med 2019;381:1632-43. [Crossref] [PubMed]
- Tian J, Chen JH, Chao SX, et al. Combined PD-1, BRAF and MEK inhibition in BRAF(V600E) colorectal cancer: a phase 2 trial. Nat Med 2023;29:458-66. [Crossref] [PubMed]
- Van Cutsem E, Taieb J, Yaeger R, et al. ANCHOR CRC: Results From a Single-Arm, Phase II Study of Encorafenib Plus Binimetinib and Cetuximab in Previously Untreated BRAF(V600E)-Mutant Metastatic Colorectal Cancer. J Clin Oncol 2023;41:2628-37. [Crossref] [PubMed]
- Hong DS, Fakih MG, Strickler JH, et al. KRAS(G12C) Inhibition with Sotorasib in Advanced Solid Tumors. N Engl J Med 2020;383:1207-17. [Crossref] [PubMed]
- Yaeger R, Weiss J, Pelster MS, et al. Adagrasib with or without Cetuximab in Colorectal Cancer with Mutated KRAS G12C. N Engl J Med 2023;388:44-54. [Crossref] [PubMed]
- Fakih MG, Kopetz S, Kuboki Y, et al. Sotorasib for previously treated colorectal cancers with KRASG12C mutation (CodeBreaK100): a prespecified analysis of a single-arm, phase 2 trial. Lancet Oncol 2022;23:115-24. [Crossref] [PubMed]
- Network NCC. NCCN Clinical Practice Guidelines in Oncology: Colon Cancer (version 1.2022). 2022.
- Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531:47-52. [Crossref] [PubMed]
- Bekaii-Saab TS, Yaeger R, Spira AI, et al. Adagrasib in Advanced Solid Tumors Harboring a KRAS(G12C) Mutation. J Clin Oncol 2023;41:4097-106. [Crossref] [PubMed]
- Strickler JH, Satake H, George TJ, et al. Sotorasib in KRAS p.G12C-Mutated Advanced Pancreatic Cancer. N Engl J Med 2023;388:33-43. [Crossref] [PubMed]
- Singhi AD, George B, Greenbowe JR, et al. Real-Time Targeted Genome Profile Analysis of Pancreatic Ductal Adenocarcinomas Identifies Genetic Alterations That Might Be Targeted With Existing Drugs or Used as Biomarkers. Gastroenterology 2019;156:2242-2253.e4. [Crossref] [PubMed]
- Philip PA, Azar I, Xiu J, et al. Molecular Characterization of KRAS Wild-type Tumors in Patients with Pancreatic Adenocarcinoma. Clin Cancer Res 2022;28:2704-14. [Crossref] [PubMed]
- Fusco MJ, Saeed-Vafa D, Carballido EM, et al. Identification of Targetable Gene Fusions and Structural Rearrangements to Foster Precision Medicine in KRAS Wild-Type Pancreatic Cancer. JCO Precis Oncol 2021;5:PO.20.00265.
- Topham JT, Tsang ES, Karasinska JM, et al. Integrative analysis of KRAS wildtype metastatic pancreatic ductal adenocarcinoma reveals mutation and expression-based similarities to cholangiocarcinoma. Nat Commun 2022;13:5941. [Crossref] [PubMed]
- Huffman BM, Ellis H, Jordan AC, et al. Emerging Role of Targeted Therapy in Metastatic Pancreatic Adenocarcinoma. Cancers (Basel) 2022;14:6223. [Crossref] [PubMed]
- Punekar SR, Velcheti V, Neel BG, et al. The current state of the art and future trends in RAS-targeted cancer therapies. Nat Rev Clin Oncol 2022;19:637-55. [Crossref] [PubMed]
- Leidner R, Sanjuan Silva N, Huang H, et al. Neoantigen T-Cell Receptor Gene Therapy in Pancreatic Cancer. N Engl J Med 2022;386:2112-9. [Crossref] [PubMed]
- Windon AL, Loaiza-Bonilla A, Jensen CE, et al. A KRAS wild type mutational status confers a survival advantage in pancreatic ductal adenocarcinoma. J Gastrointest Oncol 2018;9:1-10. [Crossref] [PubMed]
- Corcoran RB, Chabner BA. Application of Cell-free DNA Analysis to Cancer Treatment. N Engl J Med 2018;379:1754-65. [Crossref] [PubMed]
- Zill OA, Banks KC, Fairclough SR, et al. The Landscape of Actionable Genomic Alterations in Cell-Free Circulating Tumor DNA from 21,807 Advanced Cancer Patients. Clin Cancer Res 2018;24:3528-38. [Crossref] [PubMed]
- Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017;545:446-51. [Crossref] [PubMed]
- Husain H, Pavlick DC, Fendler BJ, et al. Tumor Fraction Correlates With Detection of Actionable Variants Across > 23,000 Circulating Tumor DNA Samples. JCO Precis Oncol 2022;6:e2200261. [Crossref] [PubMed]
- Bando H, Nakamura Y, Taniguchi H, et al. Effects of Metastatic Sites on Circulating Tumor DNA in Patients With Metastatic Colorectal Cancer. JCO Precis Oncol 2022;6:e2100535. [Crossref] [PubMed]
- Sivapalan L, Kocher HM, Ross-Adams H, et al. Molecular profiling of ctDNA in pancreatic cancer: Opportunities and challenges for clinical application. Pancreatology 2021;21:363-78. [Crossref] [PubMed]
- Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 2014;371:2488-98. [Crossref] [PubMed]
- Razavi P, Li BT, Brown DN, et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat Med 2019;25:1928-37. [Crossref] [PubMed]
- Leal A, van Grieken NCT, Palsgrove DN, et al. White blood cell and cell-free DNA analyses for detection of residual disease in gastric cancer. Nat Commun 2020;11:525. [Crossref] [PubMed]
- Li BT, Janku F, Jung B, et al. Ultra-deep next-generation sequencing of plasma cell-free DNA in patients with advanced lung cancers: results from the Actionable Genome Consortium. Ann Oncol 2019;30:597-603. [Crossref] [PubMed]
- Liu J, Chen X, Wang J, et al. Biological background of the genomic variations of cf-DNA in healthy individuals. Ann Oncol 2019;30:464-70. [Crossref] [PubMed]
- Mayrhofer M, De Laere B, Whitington T, et al. Cell-free DNA profiling of metastatic prostate cancer reveals microsatellite instability, structural rearrangements and clonal hematopoiesis. Genome Med 2018;10:85. [Crossref] [PubMed]
- Meador CB, Milan MSD, Hu EY, et al. High Sensitivity of Plasma Cell-Free DNA Genotyping in Cases With Evidence of Adequate Tumor Content. JCO Precis Oncol 2021;5:PO.20.00420.
- Nakamura Y, Okamoto W, Denda T, et al. Clinical Validity of Plasma-Based Genotyping for Microsatellite Instability Assessment in Advanced GI Cancers: SCRUM-Japan GOZILA Substudy. JCO Precis Oncol 2022;6:e2100383. [Crossref] [PubMed]
- Nakamura Y, Taniguchi H, Ikeda M, et al. Clinical utility of circulating tumor DNA sequencing in advanced gastrointestinal cancer: SCRUM-Japan GI-SCREEN and GOZILA studies. Nat Med 2020;26:1859-64. [Crossref] [PubMed]
- Green MF, Bell JL, Hubbard CB, et al. Implementation of a Molecular Tumor Registry to Support the Adoption of Precision Oncology Within an Academic Medical Center: The Duke University Experience. JCO Precis Oncol 2021;5:PO.21.00030.
- Yaeger R, Corcoran RB. Targeting Alterations in the RAF-MEK Pathway. Cancer Discov 2019;9:329-41. [Crossref] [PubMed]
- Yaeger R, Kotani D, Mondaca S, et al. Response to Anti-EGFR Therapy in Patients with BRAF non-V600-Mutant Metastatic Colorectal Cancer. Clin Cancer Res 2019;25:7089-97. [Crossref] [PubMed]
- Guardant Health I. Guardant360 CDx Technical Information. Redwood City, CA. Available online: https://guardant360cdx.com/wp-content/uploads/guardant360-cdx-technical-information.pdf
- Therneau T, Atkinson B, Ripley B. Package 'rpart'. 2015. Available online: https://cran.r-project.org/web/packages/rpart/index.html
- Hothorn T, Lausen B. Maximally selected rank statistics in R. R News 2002;2:3-5.
- RCT. R: A language and environment for statistical computing.: R Foundation for Statistical Computing, Vienna, Austria. Available online: https://www.R-project.org/
- RT. RStudio: Integrated Development Environment for R. Rstudio, PBC, Boston, MA. Available online: http://www.rstudio.com/


