
The development and validation of pathological sections based U-Net deep learning segmentation model for the detection of esophageal mucosa and squamous cell neoplasm
Highlight box
Key findings
• This study developed U-Net based deep learning segmentation models for esophageal squamous cell carcinoma.
What is known and what is new?
• Innovations in digital pathology enable deep learning applications for pathological images, improving processing efficacy. The pathological image classification, identification and segmentation models are well established for cancers of breast, lung and skin.
• In this study, a Visual Geometry Group-based U-Net neural network was utilized to develop mucosa and tumor segmentation models. Meanwhile, tumor segmentation was additionally validated in two external datasets.
What is the implication, and what should change now?
• The models demonstrated promising mucosa and tumor segmentation capabilities. With integration of other artificial intelligence models, clinical deployment on common computers is expected. Further extensive researches into deep learning for esophageal cancer are warrant to assist in fast and accurate diagnosis.
Introduction
With a 5-year survival rate of 20%, esophageal carcinoma stands as a significant contributor to cancer-related fatalities worldwide (1). Notably, China accounts for half of the annual reported cases globally (2). The results of pathology examinations of gastroscopy biopsies and surgical specimens are the gold standard for diagnosing and staging this malignancy. It is imperative to identify tumor cells and determine the depth of wall penetration during the scrutiny of pathological sections to attain precise diagnoses (3).
The advent of digital pathology techniques, which involve the digitization of microscopic histologic slides including scanning, compression, storage, and digital display, makes the conversion of microscopic histologic slides into high resolution images possible (4,5). As an advanced technique, the acquisition of whole slide image (WSI) has become a routine practice in cancer diagnosis, which facilitates the storage of entire pathological slides in high definition and the consultations for challenging cases (6). Typically, a high-resolution WSI comprises thousands of cells and trillions of pixels to ensure intricate cellular structures identifiable. However, when dealing with lesions or regions of interest (ROIs) that are relatively small compared to the entire sections, manually scanning of whole slide under microscopy or with WSI is a time-consuming task, even for experienced pathologists (7). The available pool of expert pathologists remains insufficient to meet the substantial clinical demand for manual pathological examination. Fortunately, WSIs enable the application of artificial intelligence for pathological diagnosis, offering a potential solution to the aforementioned challenge through automated image processing (8,9).
Computer-assisted diagnosis techniques could potentially recognize informative patterns in images that transcend human limitations and boundaries. These techniques have already been implemented in medical imaging, including endoscopic imaging, computed tomography (CT) and positron emission tomography (PET) (10,11). Artificial intelligence could also assist pathologists in identifying distinctive features in WSI, with the goal of early detection and prognosis prediction. The digital pathological images deep learning technologies have yielded remarkable capabilities in detection, segmentation, and classification for carcinomas in locations such the breast, lung and skin (12-14).
The convolutional neural networks (CNNs) have already existed for decades, large biomedical datasets containing thousands of training images are still required for the generation of complex outputs. The U-Net network, initially proposed by Ronneberger et al. in 2015, has shown precise segmentations even with very few training images for biomedical domain (15). The network primarily consists of a contracting path and an expansive path, both of which have been validated for their effectiveness in biomedical image processing and segmentation across various diseases (16,17). For esophageal cancer, previous studies have demonstrated that CNN networks could achieve comparable accuracy as clinicians, especially with computed tomography-based or endoscopic datasets (18,19). However, few studies focused on the artificial intelligence based pathological images processing in esophageal carcinoma. As the only gold standard for diagnosing esophageal cancer, and determining the depth of invasion (20), this study aimed to develop deep U-Net architecture-based learning models for the segmentation of esophageal mucosa and esophageal squamous cell carcinoma (ESCC). We present this article in accordance with the TRIPOD reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-587/rc).
Methods
Datasets
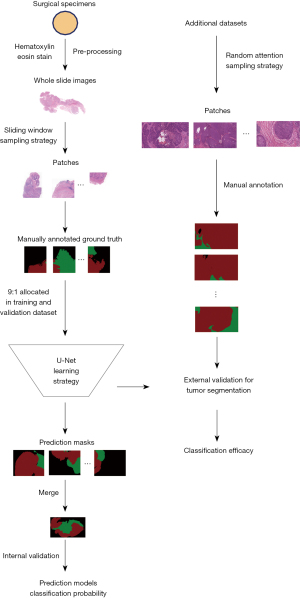
Pathological images of surgical specimen from patients with ESCC who underwent esophagectomy at the Department of Thoracic Surgery, Zhongshan Hospital, Fudan University between January 2021 and June 2021 were retrospectively reviewed as the primary dataset of the study. The inclusion criteria were: (I) age between 18 to 80 years; (II) histological diagnosis of ESCC; (III) underwent radical esophagectomy with available surgical specimens. And the exclusion criteria included: (I) history of other tumors; (II) unavailable or unqualified surgical specimens. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics review board of Zhongshan Hospital, Fudan University (No. ZS-B2021-708) and individual consent for this retrospective analysis was waived. WSIs were acquired from hematoxylin and eosin (H&E) staining of paraffin-embedded tissues, and the slides would be re-imaged if distortion, blurs, or occlusions were detected on the quality assessment. WSIs of tumor areas were allocated into tumor cohort, while WSIs of paracancerous area and normal tissue comprised mucosa cohort. Small patches were then generated based on a sliding-window sampling strategy, with patches of less than 10% ROIs eliminated. Patches in mucosa and tumor cohort were utilized for the construction of segmentation model for esophageal mucosa and ESCC, respectively. After cohort generation, patches were allocated into the training and validation sets randomly with a 9:1 split ratio. Specifically, attention was focused on the effectiveness of the tumor identification model. And therefore, two additional WSI datasets from endoscopic biopsy slides outside the Department of Pathology, Zhongshan Hospital, Fudan University, were allocated as external validation cohorts, encompassing samples from five ESCC patients each. Patches in the external validation cohorts were acquired through random window strategy sampling from WSIs.
Basic network architecture
Input patches were resized to of 512×512 pixels with matching resolution for segmentation model outputs. The basic network was U-Net structure, a convolutional network advantageous for medical image segmentation (15). It is composed of a contracting path for feature extraction and an expansive component for segmentation prediction as well (21). The Visual Geometry Group (VGG) Network was applied as the backbone for the contracting path, which consisted of repeated application of convolutions, followed by a rectified linear unit and a max pooling operation (22). And the prediction step consisted of an upsampling followed by convolutions. Each layer of the expansive component was concatenated and fused with the corresponding layer in the contracting path via skip-connection to prevent edge information loss. Besides training from scratch, pre-trained Xception, U-Net, VGG16, VGG19 and ResNet50 models were used for the transfer learning (23-25). Training epoch weights and performance were recorded to identify contributing features. The network architecture is available in Figure 1, with main hyperparameters presented in the Table 1. The deep learning process was performed on a laptop equipped with GeForce RTX3050Ti (NVIDIA) graphics processing unit and CUDA 11.0.2 for graphics processing unit (GPU) acceleration.
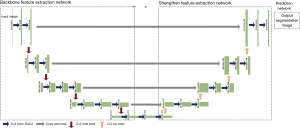
Table 1
| Hyperparameters | Values |
|---|---|
| Patch size, pixels | 512×512 |
| Batch size | 2 |
| Learning rate | 1E−4 |
| Optimizer type | Adam |
| Momentum | 0.9 |
| Epoch number | 100 |
ROIs
ROIs were manually annotated by two authors with Labelme under the assistance of Department of Pathology as the gold diagnoses of mucosa and cancer, and the generated JSON files were converted to mask map and trained on the Pytorch platform. Two ROI sets were labeled—mucosa/non-mucosa and tumor/non-tumor. Reproducibility was assessed in a blinded manner with disagreements resolved by consensus or by appealing to the third experienced thoracic surgeon. To assure the accurate diagnoses of ESCC and mucosa, all the ROIs would be verified by an experienced thoracic surgeon.
Statistical analysis
The performance of the models in predicting histological ROIs would be assessed with the following parameters: (I) the Intersection over Union (IOU); (II) the positive predictive value (PPV); (III) the true positive rate (TPR); (IV) the accuracy; (V) the specificity; (VI) the dice similarity coefficient (DSC); (VII) the area under the receiver operating characteristic curve (AUC); (VIII) the balanced F score (F1-Score) (26). Loss function is a parameter evaluating the relative differences between prediction and truth; IOU is a parameter assessing the consistency between the ground truth and the segmentation mask, IOU = TP/(TP + FP + FN); PPV is a measurement of precision, PPV = TP/(TP + FP); TPR could reflect the sensitivity, TPR = TP/(TP + FN); the accuracy = (TP + TN)/(TP + FP + FN + TN); specificity = TN/(TN + FP); DSC is a statistical measure of spatial overlap for segmentation accuracy, DSC = 2 TP/(2 TP + FP + FN); while AUC is a criteria to optimize the model hyperparameters; and the balanced F score is the harmonic mean of the sensitivity and specificity and is used to establish optimal system accuracy (TP, true positive; FN, false negative; FP, false positive; TN, true negative).
Results
Training dataset and validation datasets
A retrospective analysis was conducted on surgical samples from ten patients (mean age: 62.7 years; male: 90%). Clinical data for patients in the primary cohort and two additional cohorts is available in Table 2. A total of 30 pairs of carcinomas and para-carcinomas tissues were obtained from surgical specimens and subsequently underwent H&E staining. Following WSI pre-processing, 292 and 239 patches were derived for the mucosa and tumor cohort with a sliding window sampling approach, respectively. Meanwhile, the two additional datasets primarily featured tumor-dominant WSIs, from which 59 and 48 patches were generated for external validation cohort 1 and validation cohort 2 following random attention sampling, respectively. An overview of the training and validation process is depicted in the Figure 2.
Table 2
| Characteristics | Main cohort | Additional cohort 1 | Additional cohort 2 |
|---|---|---|---|
| Patients | 10 | 5 | 5 |
| Age (years) | 62.7±5.0 | 67.4±6.7 | 57.4±7.2 |
| Male | 9 [90] | 5 [100] | 4 [80] |
| Tumor location | |||
| Middle | 8 [80] | 5 [100] | 3 [60] |
| Lower | 2 [20] | 0 [0] | 2 [40] |
| Clinical stage | |||
| II | 5 [50] | 5 [100] | 4 [80] |
| III | 5 [50] | 0 [0] | 1 [20] |
| Histological grade | |||
| 2 | 9 [90] | 3 [60] | 4 [80] |
| 3 | 1 [10] | 2 [40] | 1 [20] |
Data are shown as mean ± SD or n [%]. ESCC, esophageal squamous cell carcinoma; SD, standard deviation.
Performance in the deep learning for mucosa
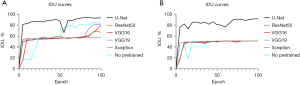
There were 262 and 30 patches allocated in the training and internal validation cohort for the mucosa segmentation task. The performance of models exhibited variability when different pretrained models were applied, however, all models consistently demonstrated robust discriminatory capability between mucosa and non-mucosa composition after 100 epochs of training (Table 3). The ascension of IOU in the training is shown in Figure 3A. Notably, the trained U-Net model exhibited the highest potential, achieving an IOU of 93.81%, accuracy of 96.96%, PPV of 96.45%, TPR of 96.65%, specificity of 98.41%, DSC of 96.81%, AUC of 96.11% and F1-Score of 96.55% in the internal validation. Besides, the transfer learning VGG19 (IOU =82.52%) model and model trained from scratch (IOU =84.70%) also demonstrated commendable identification abilities, with most parameters basically reaching 90%.
Table 3
| Models | IOU (%) | Accuracy (%) | PPV (%) | TPR (%) | Specificity (%) | DSC (%) | AUC (%) | F1-Score (%) |
|---|---|---|---|---|---|---|---|---|
| U-Net | 93.81 | 96.96 | 96.45 | 96.65 | 98.41 | 96.81 | 96.11 | 96.55 |
| VGG16 | 75.69 | 82.75 | 87.47 | 89.87 | 89.55 | 86.16 | 82.62 | 88.65 |
| VGG19 | 82.52 | 90.52 | 89.35 | 90.33 | 94.77 | 90.42 | 88.65 | 89.84 |
| ResNet50 | 68.23 | 75.74 | 84.52 | 87.31 | 84.85 | 81.12 | 76.54 | 85.89 |
| Xception | 58.28 | 67.00 | 78.99 | 81.74 | 78.88 | 73.64 | 68.58 | 80.34 |
| No pretrained | 84.70 | 90.75 | 91.79 | 92.70 | 94.86 | 91.72 | 89.78 | 92.24 |
IOU, Intersection over Union; PPV, positive predictive value; TPR, true positive rate; DSC, dice similarity coefficient; AUC, area under the receiver operating characteristic curve; F1-Score, balanced F score.
Performance in the deep learning for tumor
In the tumor segmentation, the training and internal validation cohorts consisted of 215 and 24 patches respectively. The IOU curves of models are displayed in the Figure 3B and the performance of models in the internal validation is summarized in Table 4. Except the U-Net model, most models performed poorly, with IOU not exceeding 52%. The U-Net based model outperformed all other models, achieving an IOU of 91.95%, accuracy of 95.90%, PPV of 95.62%, TPR of 95.71%, specificity of 97.88%, DSC of 95.81%, AUC of 94.92% and F1-Score of 95.67%.
Table 4
| Models | IOU (%) | Accuracy (%) | PPV (%) | TPR (%) | Specificity (%) | DSC (%) | AUC (%) | F1-Score (%) |
|---|---|---|---|---|---|---|---|---|
| U-Net | 91.95 | 95.90 | 95.62 | 95.71 | 97.88 | 95.81 | 94.92 | 95.67 |
| VGG16 | 51.01 | 65.71 | 51.75 | 69.51 | 73.81 | 67.56 | 62.41 | 59.33 |
| VGG19 | 51.05 | 65.72 | 51.73 | 69.58 | 73.79 | 67.59 | 62.42 | 59.34 |
| ResNet50 | 51.16 | 65.80 | 51.80 | 69.69 | 73.84 | 67.69 | 62.50 | 59.43 |
| Xception | 51.11 | 65.71 | 51.76 | 69.70 | 73.75 | 67.65 | 62.43 | 59.41 |
| No pretrained | 51.15 | 65.89 | 51.69 | 69.57 | 73.93 | 67.68 | 62.54 | 59.31 |
IOU, Intersection over Union; PPV, positive predictive value; TPR, true positive rate; DSC, dice similarity coefficient; AUC, area under the receiver operating characteristic curve; F1-Score, balanced F score.
External validation for tumor segmentation
Given the clinical value of tumor segmentation, the performance of models, especially the U-Net based model, were validated on two additional datasets.
Dataset 1: the results of external validation for dataset 1 are summarized in Table 5. As expected, U-Net outperformed other models. The VGG16, VGG19, ResNet50, Xception and scratch-based models yielded IOU of 25%, while U-Net model attained 59.86% for overall IOU. Notably, the resolution capability of U-Net model for tumor segmentation was specifically investigated, yielding 79.00% for IOU, 86.00% for accuracy, 90.00% for PPV, 90.66% for TPR, 92.21% for specificity, 88.27% for DSC, 85.61% for AUC, and 90.33% for F1-Score.
Table 5
| Models | IOU (%) | Accuracy (%) | PPV (%) | TPR (%) | Specificity (%) | DSC (%) | AUC (%) | F1-Score (%) |
|---|---|---|---|---|---|---|---|---|
| U-Net for tumors | 79.00 | 86.00 | 90.00 | 90.66 | 92.21 | 88.27 | 85.61 | 90.33 |
| U-Net | 59.86 | 72.74 | 76.03 | 77.17 | 83.80 | 74.89 | 71.83 | 76.60 |
| VGG16 | 25.09 | 51.53 | 38.36 | 32.84 | 70.73 | 40.12 | 47.91 | 35.39 |
| VGG19 | 25.00 | 52.28 | 36.08 | 32.39 | 70.47 | 40.00 | 47.73 | 34.14 |
| ResNet50 | 25.61 | 52.45 | 37.41 | 33.35 | 70.76 | 40.78 | 48.19 | 35.27 |
| Xception | 25.64 | 54.21 | 33.86 | 32.73 | 70.84 | 40.82 | 48.24 | 33.28 |
| No pretrained | 25.68 | 52.44 | 37.64 | 33.48 | 70.80 | 40.87 | 48.24 | 35.44 |
IOU, Intersection over Union; PPV, positive predictive value; TPR, true positive rate; DSC, dice similarity coefficient; AUC, area under the receiver operating characteristic curve; F1-Score, balanced F score.
Dataset 2: in the dataset 2, similar result of external validation as dataset 1 was achieved (Table 6). The performance of 5 models other than U-Net was far from satisfactory. Interestingly, the prediction ability of the whole model in the dataset 2 (U-Net achieved 50.88% for IOU, 68.03% for accuracy, 59.02% for PPV, 66.87% for TPR, 78.48% for specificity, 67.44% for DSC, 64.68% for AUC, and 62.70% for F1-Score) might be not as good as that in dataset 1, however, the differentiation ability of tumor tissue surpassed the dataset 1 (IOU of 89.00%, accuracy of 91.00%, PPV of 97.00%, TPR of 97.59%, specificity of 94.78%, DSC of 94.18%, AUC of 91.89% and F1-Score of 97.29%).
Table 6
| Models | IOU (%) | Accuracy (%) | PPV (%) | TPR (%) | Specificity (%) | DSC (%) | AUC (%) | F1-Score (%) |
|---|---|---|---|---|---|---|---|---|
| U-Net for tumors | 89.00 | 91.00 | 97.00 | 97.59 | 94.78 | 94.18 | 91.89 | 97.29 |
| U-Net | 50.88 | 68.03 | 59.02 | 66.87 | 78.48 | 67.44 | 64.68 | 62.70 |
| VGG16 | 5.00 | 34.80 | 9.59 | 5.52 | 60.05 | 9.52 | 32.53 | 7.00 |
| VGG19 | 5.55 | 36.31 | 7.59 | 6.15 | 56.23 | 10.52 | 30.89 | 6.79 |
| ResNet50 | 6.06 | 36.86 | 9.12 | 6.76 | 58.19 | 11.43 | 32.12 | 7.77 |
| Xception | 10.32 | 44.37 | 14.59 | 11.85 | 64.42 | 18.71 | 37.37 | 13.08 |
| No pretrained | 5.96 | 36.34 | 10.08 | 6.65 | 59.49 | 11.25 | 32.72 | 8.02 |
IOU, Intersection over Union; PPV, positive predictive value; TPR, true positive rate; DSC, dice similarity coefficient; AUC, area under the receiver operating characteristic curve; F1-Score, balanced F score.
Discussion
In this article, a rigorous development and validation of novel deep learning models for segmenting ESCC pathology were presented. And the model based on the U-Net pretrained parameters has demonstrated outstanding performance for mucosa and tumor segmentation in both internal and external validation. To the best of our knowledge, this is a pioneering study exploring deep learning methods for pathological images segmentation of esophageal carcinoma.
The rise and widespread adoption of digital image system and WSI have enabled features extraction and quantitative histomorphometry analysis. The implementation of deep learning based artificial intelligence assists on various tasks including diagnosis, grading and subtyping for carcinomas located in breast, lung and digestive tract (27). Based on CNNs, deep learning can ideally deal with the complex digital pathological image analysis tasks, and it could serve as a promising frontier. Previous studies have documented the utilization of CNNs in the classification for esophageal carcinoma H&E-stained images (7,28). In addition to classification, CNNs are also implemented in tumor image segmentation (29). In order to achieve precise segmentation, various networks structures have been proposed (21,30). However, current deep learning techniques for tumor segmentation primarily focused on morphological images including ultrasound (US), X-ray, CT, magnetic resonance imaging (MRI), and PET/CT, with only a limited emphasis on pathological images of certain carcinomas. This article delves into the viability of integrating deep learning techniques with pathological WSIs, widely regarded as the benchmark for tumor diagnosis. On the other hand, the development of traditional CNN network demands a sizable image cohort to ensure efficacy, while the U-Net network has demonstrated the ability to segment biomedical images effectively with minimal training data. To ensure the effectiveness of models with limited sample size, we applied the U-Net segmentation model in our study and achieved satisfactory results.
In this study, the U-Net framework was used for the segmentation learning, with the VGG net serving as the backbone. In the absence of specific threshold values, we basically assumed a threshold of 80% for parameters. Notably, apart from learning from scratch, transfer learning, a common technique in artificial intelligence, was employed to explore compatible segmentation models. In artificial intelligence community, finely tuned pre-trained models through transfer learning are utilized to reduce training time and potentially achieve better results. The IOU curves of transfer learning models reached a performance plateau in fewer epochs compared to models learned from scratch. This might be attributed to inherent differences between digital pathological images and images in ImageNet datasets. Moreover, compared to previous network structures, the U-Net model demonstrated several advantages. The corresponding layers in the expansive path and contracting path were seamed in order to ensure the sufficient features were extracted from limited images, as a result, a large number of feature channels in the upsampling part allowed the network to propagate characteristics information to higher resolution layers (15). Besides, the adoption of a tiling strategy enables the prediction of border region in large images without consuming excessive GPU memory. Therefore, it was expected that the U-Net based transfer learning model would exhibit superior performance in both mucosa and tumor segmentation in esophageal carcinoma, which was aligned with the findings of this study.
The current study has several strengths. Firstly, the employed networks, especially the U-Net transfer learning model, appeared capable of segmenting ESCC WSIs, generating accurate ROI segmentation masks with clear borderline. This indicates its potential clinical application in tumor staging and sections screening for surgical specimen. Even with a small training cohort, models demonstrated acceptable efficacy on external validation. Secondly, this study is the first exploration of the deep learning segmentation for ESCC pathology images, establishing a foundation to guide future researches. The mucosa segmentation model could aid in the tumor stage of ESCC, and tumor segmentation models may help pathologists rapidly screen slides and identify concerning areas. Moreover, previous studies have also demonstrated systems detecting neoplasia in endoscopic images to guide biopsy, these segmentation models could facilitate biopsy diagnosis (19,31). The combination of multiple deep learning segmentation systems could assist diagnosis and clinical decision, especially the feasibility of minimal invasive surgery. Overall, these strengths highlight the significance of this study and its potential implications for improving the diagnosis and management of ESCC.
This study has several limitations as well. In the first place, despite appealing results, the cohort was relatively small. Cancer heterogeneity means that limited sample size might affect the generalizability of the model, potentially contributing to poor segmentation in the external validation. Expanding the cohort, particularly with clinically studied patients, would improve the robustness of models. Besides, even though the VGG based U-Net network structure is highly regarded for medical images segmentation, the efficacy of other network architectures warrant exploration, and in further studies, comparisons should be made between the segmentation ability of different network structures. Based on limitations above, the model has not been applied in clinical practice yet. The model could be expected to be deployed on common computers and combined with other artificial intelligence models in clinical applications, while mature models developed with more extensive, diverse data are prerequisite for the clinical practice.
Conclusions
In conclusion, a deep learning framework for surgical pathology segmentation in ESCC was successfully developed and validated using a VGG based U-Net network in this study. The model based on U-Net pretraining weights offered a promising performance in contouring accurately mucosa and tumor in particular. These results hold potential for advancing quantitative histopathology diagnosis methodologies for patients with ESCC after surgery. Importantly, these segmentation models could be deployed on common computers or even personal laptops embedded with medical image equipment, offering the capabilities to highlight the ROIs for ESCC, thereby enhancing clinical decision making. Further studies will be needed to train these models in a comprehensive clinical WSI databases to distill the complex heterogeneity encountered in real-world clinical practice.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (No. 81400681), the China Postdoctoral Science Foundation Grant (No. 2018M631394), and the Science and Technology Innovation Plan of Shanghai Science and Technology Commission (No. 22Y11907200). The funding agencies had no role in study design, collection and analyses of data, decision to publish, or manuscript preparation.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-587/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-587/dss
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-587/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-587/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics review board of Zhongshan Hospital, Fudan University (No. ZS-B2021-708) and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin 2022;72:7-33. [Crossref] [PubMed]
- Li J, Chen B, Wang X, et al. Prognosis of patients with esophageal squamous cell carcinoma undergoing surgery versus no surgery after neoadjuvant chemoradiotherapy: a retrospective cohort study. J Gastrointest Oncol 2022;13:903-11. [Crossref] [PubMed]
- Waters JK, Reznik SI. Update on Management of Squamous Cell Esophageal Cancer. Curr Oncol Rep 2022;24:375-85. [Crossref] [PubMed]
- Niazi MKK, Parwani AV, Gurcan MN. Digital pathology and artificial intelligence. Lancet Oncol 2019;20:e253-61. [Crossref] [PubMed]
- Bashshur RL, Krupinski EA, Weinstein RS, et al. The Empirical Foundations of Telepathology: Evidence of Feasibility and Intermediate Effects. Telemed J E Health 2017;23:155-91. [Crossref] [PubMed]
- Jahn SW, Plass M, Moinfar F. Digital Pathology: Advantages, Limitations and Emerging Perspectives. J Clin Med 2020;9:3697. [Crossref] [PubMed]
- Tomita N, Abdollahi B, Wei J, et al. Attention-Based Deep Neural Networks for Detection of Cancerous and Precancerous Esophagus Tissue on Histopathological Slides. JAMA Netw Open 2019;2:e1914645. [Crossref] [PubMed]
- Madabhushi A, Lee G. Image analysis and machine learning in digital pathology: Challenges and opportunities. Med Image Anal 2016;33:170-5. [Crossref] [PubMed]
- Zhu J, Liu M, Li X. Progress on deep learning in digital pathology of breast cancer: a narrative review. Gland Surg 2022;11:751-66. [Crossref] [PubMed]
- van der Sommen F, Curvers WL, Nagengast WB. Novel Developments in Endoscopic Mucosal Imaging. Gastroenterology 2018;154:1876-86. [Crossref] [PubMed]
- Avanzo M, Stancanello J, Pirrone G, et al. Radiomics and deep learning in lung cancer. Strahlenther Onkol 2020;196:879-87. [Crossref] [PubMed]
- Huang B, Tian S, Zhan N, et al. Accurate diagnosis and prognosis prediction of gastric cancer using deep learning on digital pathological images: A retrospective multicentre study. EBioMedicine 2021;73:103631. [Crossref] [PubMed]
- Kawahara D, Murakami Y, Tani S, et al. A prediction model for pathological findings after neoadjuvant chemoradiotherapy for resectable locally advanced esophageal squamous cell carcinoma based on endoscopic images using deep learning. Br J Radiol 2022;95:20210934. [Crossref] [PubMed]
- Li F, Yang Y, Wei Y, et al. Deep learning-based predictive biomarker of pathological complete response to neoadjuvant chemotherapy from histological images in breast cancer. J Transl Med 2021;19:348. [Crossref] [PubMed]
- Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab N, Hornegger J, Wells WM, et al. editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Cham: Springer International Publishing; 2015:234-41.
- Swiderska-Chadaj Z, Pinckaers H, van Rijthoven M, et al. Learning to detect lymphocytes in immunohistochemistry with deep learning. Med Image Anal 2019;58:101547. [Crossref] [PubMed]
- Zhou T, Tan T, Pan X, et al. Fully automatic deep learning trained on limited data for carotid artery segmentation from large image volumes. Quant Imaging Med Surg 2021;11:67-83. [Crossref] [PubMed]
- Hu Y, Xie C, Yang H, et al. Computed tomography-based deep-learning prediction of neoadjuvant chemoradiotherapy treatment response in esophageal squamous cell carcinoma. Radiother Oncol 2021;154:6-13. [Crossref] [PubMed]
- de Groof AJ, Struyvenberg MR, van der Putten J, et al. Deep-Learning System Detects Neoplasia in Patients With Barrett's Esophagus With Higher Accuracy Than Endoscopists in a Multistep Training and Validation Study With Benchmarking. Gastroenterology 2020;158:915-929.e4. [Crossref] [PubMed]
- Mönig S, Chevallay M, Niclauss N, et al. Early esophageal cancer: the significance of surgery, endoscopy, and chemoradiation. Ann N Y Acad Sci 2018;1434:115-23. [Crossref] [PubMed]
- Jiang HY, Diao ZS, Yao YD. Deep learning techniques for tumor segmentation: a review. Journal of Supercomputing 2022;78:1807-51. [Crossref]
- Chen W, Wang WP, Liu L, et al. New ideas and trends in deep multimodal content understanding: a review. Neurocomputing 2021;426:195-215. [Crossref]
- Chollet F, IEEE. Xception: Deep Learning with Depthwise Separable Convolutions. 30th IEEE Conference on Computer Vision and Pattern Recognition (CVPR 2017) 2017:1800-7.
- Szegedy C, Ioffe S, Vanhoucke V, et al. Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. Thirty-first AAAI Conference on Artificial Intelligence 2017:4278-84.
- Deng J, Dong W, Socher R, et al. ImageNet: A Large-Scale Hierarchical Image Database. CVPR: 2009 IEEE Conference on Computer Vision and Pattern Recognition 2009:248-55.
- Taha AA, Hanbury A. Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool. BMC Med Imaging 2015;15:29. [Crossref] [PubMed]
- Jiang Y, Yang M, Wang S, et al. Emerging role of deep learning-based artificial intelligence in tumor pathology. Cancer Commun (Lond) 2020;40:154-66. [Crossref] [PubMed]
- Beuque M, Martin-Lorenzo M, Balluff B, et al. Machine learning for grading and prognosis of esophageal dysplasia using mass spectrometry and histological imaging. Comput Biol Med 2021;138:104918. [Crossref] [PubMed]
- Xu J, Luo X, Wang G, et al. A Deep Convolutional Neural Network for segmenting and classifying epithelial and stromal regions in histopathological images. Neurocomputing (Amst) 2016;191:214-23. [Crossref] [PubMed]
- Rezaeijo SM, Jafarpoor Nesheli S, Fatan Serj M, et al. Segmentation of the prostate, its zones, anterior fibromuscular stroma, and urethra on the MRIs and multimodality image fusion using U-Net model. Quant Imaging Med Surg 2022;12:4786-804. [Crossref] [PubMed]
- Islam MM, Poly TN, Walther BA, et al. Deep Learning for the Diagnosis of Esophageal Cancer in Endoscopic Images: A Systematic Review and Meta-Analysis. Cancers (Basel) 2022;14:5996. [Crossref] [PubMed]


