
Cadherin dysregulation in gastric cancer: insights into gene expression, pathways, and prognosis
Highlight box
Key findings
• Cadherin dysregulation plays a significant role in gastric cancer (GC) development and progression, with potential as biomarkers and therapeutic targets.
What is known and what is new?
• Dysregulation of cadherin expression and function has been associated with various gastric diseases, particularly GC. However, the specific mechanisms and prognostic significance of cadherin dysregulation in GC remain unclear.
• In this study, the expression, mutational status, and prognostic potential of cadherin family genes in stomach adenocarcinoma through bioinformatics analysis were investigated.
What is the implication, and what should change now?
• This study provides comprehensive insights into the dysregulation of cadherin genes in GC and their impact on gene expression, pathways, and prognosis. These findings highlight the potential role of cadherin genes as prognostic markers and therapeutic targets in GC.
Introduction
Stomach adenocarcinoma (STAD) is the fifth most common cancer globally and the third leading cause of cancer-related deaths (1). The incidence of STAD varies geographically, with the highest rates seen in Eastern Asia and Latin America (2). Risk factors for STAD include infection with the bacterium Helicobacter pylori, a diet high in salt and processed foods, smoking, and a family history of the disease (3-7). Clinical symptoms of STAD may include abdominal pain, nausea, vomiting, weight loss, and anemia (8,9). The current treatment options for STAD include surgery, chemotherapy, targeted therapy, immunotherapy and radiation therapy (10-12). However, the effectiveness of these treatments depends on the stage of the disease at diagnosis, and the prognosis for patients with advanced stage disease remains poor.
Cadherin family genes are a group of transmembrane proteins that play crucial roles in cell-cell adhesion, tissue morphogenesis, and embryonic development (13-15). Cadherins are expressed in a wide variety of tissues, including the stomach, and are known to mediate cell-cell interactions that are essential for the maintenance of tissue architecture (16). Dysregulation of cadherin gene expression and function has been implicated in the development and progression of many cancers, including STAD (17-22). Aberrant expression or loss of function of cadherins can result in altered cell adhesion, migration, and invasion, leading to increased tumor cell dissemination and metastasis. Cadherins are known to promote as well as inhibit cancer growth and can function both as oncogenes and tumor suppressors (14,23,24). For example, E-cadherin, a major cell-cell-adhesion molecule, functions as a tumor suppressor and its downregulation results in metastasis of tumor cells (24). Further, expression of wild-type E-cadherin was shown to significantly inhibit the growth of colorectal tumor cell line (25). In contrast, proteolysis of E-cadherin generates fragments that promote tumor growth, survival, and motility, indicating cleavage of E-cadherin transforms this tumor suppressor into an oncogenic factor in certain cancer types (23). Such dual roles for other cadherins in cancer have been reviewed in detail elsewhere (14). Additionally, cadherin family genes are attractive targets for cancer therapy, as they are important in tumor progression and are often differentially expressed in cancer cells compared to normal cells (26). Thus, studying the expression, mutational status, and prognostic potential role of cadherin family genes in STAD is critical for identifying new biomarkers and therapeutic targets that may improve patient outcomes.
Several studies investigated the relationship between dysregulation of cadherin gene expression and STAD (27-30). For example, Long et al. reported that high CDH17 expression was associated with more advanced stages (III–IV vs. I–II), higher histologic grades (3–4 vs. 1–2), increased invasion grades (T3–4 vs. T1–2), and lymph node metastasis (positive vs. negative) in GC (27). In other studies, Hansford et al. evaluated the risks of GC in individuals with germline mutations in the E-cadherin (CDH1) gene. They identified 31 different pathogenic CDH1 mutations among 34 patients with GC. Based on their findings, the cumulative lifetime incidence of GC in individuals with germline CDH1 mutations at the age of 80 was estimated to be 70% for males and 56% for females (28). Donner et al. reported that mutations in the CDH1 gene are found in 30% of hereditary diffuse gastric cancer (HDGC) families and a germline truncating mutation in the gene encoding α-E-catenin (CTNNA1) was discovered in some families with HDGC (29). Lobo et al. also found that germline variants in the CTNNA1 gene are significantly associated with HDGC (30). In addition, several studies have explored the relationship between cadherin gene expression and patient survival, with different results. Although some studies have reported that decreased expression of cadherin genes is associated with poor outcomes (31,32), others have found poorer outcomes in case of increased expression of cadherin genes (27). Overall, the existing literature suggests that cadherin family genes are key players in the development and progression of STAD, and further investigation is warranted to fully understand their clinical significance.
Despite the existing literature on cadherin family genes in STAD, there are still several gaps in our knowledge that need to be addressed. For instance, it is not clear how many cadherin family members are associated with the pathogenesis of STAD, which cadherin family member has the greatest effect on development of STAD, and what is the molecular mechanism through which cadherin genes are involved in STAD progression. To fill these gaps, our study aimed to investigate the expression, mutational status, and prognostic role of cadherin family genes in STAD using bioinformatics analysis. Specifically, we analyzed differential expression, mutations and copy number variations (CNVs), survival rate, microRNA (miRNA) targeting cadherin, and immune cell infiltration in STAD tissue samples to identify potential biomarkers and therapeutic targets for this disease, as well as contribute to a better understanding of the function of cadherin family genes in this disease. We present this article in accordance with the STREGA reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-700/rc).
Methods
Cadherin family genes and estimating their differential expression
The cadherin family of genes were collected using the HGNC portal (https://www.genenames.org/data/genegroup/#!/group/16). This superfamily consists of three subfamilies, including major cadherins, protocadherins, and cadherin-related. The first 2 subfamilies are further sub-divided into specific groups. The differential expression of cadherins between STAD and normal tissues were obtained from Gene Expression Profiling Interactive Analysis 2 (GEPIA2; http://gepia2.cancer-pku.cn/) (33), which applies two different methods [analysis of variance (ANOVA) and limma] for the differential expression calculation of genes, through the use of The Cancer Genome Atlas (TCGA) (34) and Genotype Tissue Expression (GTEx) (35) RNA-seq data. We selected cadherins that were significantly differentially expressed between STAD and normal samples with a |log2 fold change (FC) ≥1| and Benjamini and Hochberg (B&H) corrected P value <0.05. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Cadherin-correlated genes and functional analysis
To understand the functions of the selected cadherins, we identified cadherin-coexpressed genes using GEPIA2, which considers tumor RNA sequencing expression data from the TCGA project (34) and analyzes them using standard processing pipelines. The ‘Similar Genes Detection’ program (http://gepia2.cancer-pku.cn/#similar) was used to obtain the correlated genes. This tool provided the top 1,000 genes with a similar expression pattern to the cadherin genes along with their correlation coefficient values calculated by Pearson correlation method. The correlated genes were used for performing functional analysis including Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology biological process (GO-BP) enrichment through WebGestalt (http://www.webgestalt.org/) (36). The annotations with B&H corrected P<0.1 were considered statistically significant. Hiplot (https://hiplot.cn/) (37) was used to visualize the pathway and GO-BP results.
Analysis of mutations and CNVs
Genomic aberrations, including single nucleotide variants, CNVs, and structural variants for differentially expressed cadherin genes were analyzed using cBioPortal (https://www.cbioportal.org/) (38). This portal provides a web resource for exploring, visualizing, and analyzing multidimensional cancer genomics data and uses TCGA as one of the sources for cancer omics data analysis. We used all TCGA GC samples (n=440) for the analysis of genomic aberrations. Of these, 436 samples have mutation and structural variant data, whereas 438 samples have copy number alteration data. Furthermore, muTarget (https://www.mutarget.com/) (39) was used to identify genes that are differentially expressed between cadherin-mutated and cadherin-wildtype GC samples. muTarget combines RNA-sequencing and mutation data to identify gene expression changes related to a gene mutation. For GC, there were 372 samples that had both messenger RNA (mRNA) expression and mutation data in muTarget. The tool was queried with each selected cadherin with the following filters: ‘Tumor type(s): Gastric cancer’ ‘Mutation type: All somatic mutations’, ‘P value cutoff: 0.01’, ‘Fold change cutoff: 2’, ‘FDR cutoff: 5%’, and ‘Exclude genes with a mean expression below 100’ to identify genes that are differentially expressed between cadherin-mutated and cadherin-wildtype GC samples. The cadherins mutated in less than 10 patient samples were not considered for the analysis.
Survival analysis
We used the Kaplan-Meier Plotter (https://kmplot.com/) (40,41) and the University of Alabama at Birmingham Cancer data analysis Portal (UALCAN; http://ualcan.path.uab.edu/index.html) (42) databases to assess the association between differentially expressed cadherins and overall survival (OS) of GC patients. Both resources integrate survival and mRNA expression data from TCGA. Additionally, UALCAN calculates effect of mRNA expression and race or gender of patients on their survival. The associations with a P<0.05 were considered statistically significant.
miRNA analysis
We used the miRDB database (https://mirdb.org/) (43) to predict miRNAs targeting the differentially expressed cadherins. In miRDB, the targets have been predicted by MirTarget tool, which was developed by analyzing thousands of miRNA-target interactions from high-throughput sequencing experiments. The experimentally verified interactions between miRNA-cadherin were obtained from the miRTarBase (https://mirtarbase.cuhk.edu.cn) (44). Furthermore, differentially expressed miRNAs between GC and normal samples were identified based on TCGA-miRNA seq data using dbDEMC database v3.0 (https://www.biosino.org/dbDEMC/) (45). This database uses the limma method (46) to identify differential expression of miRNAs between cancer and normal samples. The miRNAs with an adjusted P<0.05 (B&H correction) and an absolute FC of 1.5 were considered significant. A network between the selected cadherins and corresponding miRNAs was constructed using Cytoscape 3.9.1 (47). Only differentially expressed miRNAs were considered for network construction. Furthermore, the correlation between miRNA expression and their corresponding cadherin targets was assessed by Pearson correlation method.
Immune cell infiltration analysis
We performed immune cell infiltration analysis using ImmuCellAI portal (http://bioinfo.life.hust.edu.cn/ImmuCellAI) (48). TCGA RNA-seq data (TPM values) for stomach cancer and normal patients were uploaded onto ImmuCellAI and abundance of 24 different immune cell types were estimated in each group using the single-sample gene set enrichment analysis (ssGSEA) algorithm. Then, the significance of differential immune cells infiltration between cancer and normal tissues were calculated using unpaired t-test, and values with P<0.05 were considered significant. Further, correlation between the expression of cadherin genes and abundance of immune cell types were derived using Pearson correlation method. The correlation analysis was performed using corrplot R package (https://github.com/taiyun/corrplot) and the correlation R values with P<0.05 were considered significant.
Results
Cadherin genes are differentially expressed between STAD and normal samples
We used the HGNC portal to collect a total of 125 cadherin family of genes. These belonged to seven different groups, namely 7D cadherins (n=2), cadherin-related (n=17), CELSR cadherins (n=3), clustered protocadherins (n=64), desmosomal cadherins (n=7), major cadherins (n=2), non-clustered protocadherins (n=12), type I classical (n=5), and type II classical cadherins (n=13). Further, we used RNA-seq data to identify cadherins that showed significant differential expression between stomach cancer (TCGA data, n=408) and normal (TCGA + GTEx data, n=211) samples. Of 125 cadherins, 16 genes were identified to be significantly different in their expression with |log2FC >1| and false discovery rate (FDR)-P<0.05 (Table 1). Among these, 4 belonged to the cadherin-related group, 3 each belonged to type I classical cadherins and non-clustered protocadherins, 2 belonged to desmosomal cadherins, and 1 each belonged to the type II classical, major, 7D cadherin, and clustered protocadherin groups. It is noteworthy that all the cadherins except CDH2 were upregulated in GC compared to normal controls (Table 1). Furthermore, although PCDHGA10, PCDH17, and CDH2 were significantly different between cancer and normal samples, their relative expression was low [median transcripts per million (TPM) <5] in both these sample groups (Figure 1A). CDH17 was highly upregulated in STAD, with a nearly 5-fold difference (log2 scale) between the tumor and normal samples (Figure 1B). The other cadherins that were highly upregulated with a log2FC >2 included CDHR5, DSC2, DSG2, CDH3, and CDH11. The only downregulated cadherin, CDH2 was found to be 1.2-fold lower in cancer patients compared to normal controls (Table 1 and Figure 1B).
Table 1
| Gene symbol | Gene name | Cadherin group | Log2 fold change (limma) | Adjusted P value (limma) |
|---|---|---|---|---|
| CDH1 | Cadherin 1 | Type I classical | 1.598 | 1.12E−48 |
| CDH11 | Cadherin 11 | Type II classical | 2.281 | 2.44E−57 |
| CDH13 | Cadherin 13 | Major cadherin | 1.719 | 1.95E−46 |
| CDH17 | Cadherin 17 | 7D cadherin | 4.908 | 3.79E−74 |
| CDH2 | Cadherin 2 | Type I classical | −1.225 | 3.84E−32 |
| CDH3 | Cadherin 3 | Type I classical | 3.137 | 2.19E−97 |
| CDHR2 | Cadherin related family member 2 | Cadherin related | 1.614 | 7.73E−14 |
| CDHR5 | Cadherin related family member 5 | Cadherin related | 3.544 | 4.82E−67 |
| CLSTN1 | Calsyntenin 1 | Cadherin related | 1.437 | 2.91E−66 |
| DSC2 | Desmocollin 2 | Desmosomal | 2.008 | 2.96E−34 |
| DSG2 | Desmoglein 2 | Desmosomal | 2.645 | 1.5E−100 |
| FAT1 | FAT atypical cadherin 1 | Cadherin related | 1.953 | 3.6E−98 |
| PCDH1 | Protocadherin 1 | Non-clustered protocadherin | 1.518 | 4.43E−46 |
| PCDH17 | Protocadherin 17 | Non-clustered protocadherin | 1.192 | 7.76E−60 |
| PCDH18 | Protocadherin 18 | Non-clustered protocadherin | 1.01 | 7.88E−30 |
| PCDHGA10 | Protocadherin gamma subfamily A, 10 | Clustered protocadherin | 1.079 | 3.39E−34 |
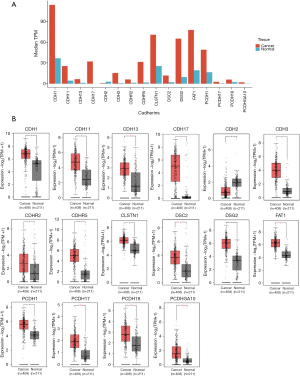
Cadherin-correlated genes in GC are associated with adhesion and cancer-related pathways
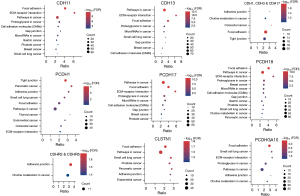
Genes with similar expression pattern are commonly used to infer the functional roles of their partners. Hence, we first identified genes that are correlated (Pearson correlation coefficient ≥0.25) with the differentially expressed cadherins identified by us in GC compared to normal samples, and then used those correlated genes for functional enrichment analysis. As expected, the correlated genes corresponding to most of the cadherins were involved in cancer, adhesion, extracellular matrix (ECM)-receptor interaction, or junction related pathways (all pathways were significant with FDR P<0.1). Figure 2 shows selected pathways enriched by the genes correlated with differential cadherins. A complete list of pathways can be found in Supplementary file available at https://cdn.amegroups.cn/static/public/jgo-23-700-1.xlsx. CDH13- followed by PCDH1-correlated genes were enriched in most KEGG pathways (n=66 and 65, respectively). Focal adhesion and ECM-receptor interaction were the top 10 pathways for CDH11-, CDH13-, PCDH17-, PCDH18-, and PCDHGA10-correlated genes, indicating that these genes are directly involved in adhesion or junction related functions. CDH1-correlated genes were mainly involved in endocytosis, and autophagy/mitophagy related pathways. CDH17- and CDHR2-correlated genes were involved in metabolic pathways, such as tricarboxylic acid cycle (TCA) cycle, fructose and mannose metabolism, and propanoate metabolism. CDHR5-correlated genes were primarily enriched in fat digestion and glucose metabolism pathways. FAT1-correlated genes were involved in apoptosis, RNA transport, and splicing pathways. The GO-BP analysis results were in similar lines to that of pathway enrichment (all annotations were significant with FDR P<0.1). For instance, CDH1-correlated genes were enriched for transport and vesicular organization related BPs. Similarly, CDH11- and CDH13-correlated genes were involved in adhesion and tissue migration-related processes. Supplementary file available at https://cdn.amegroups.cn/static/public/jgo-23-700-2.xlsx represents the list of GO-BPs significantly enriched by cadherin-correlated genes.
Cadherins are altered in GC patients and affect the expression of other genes
In TCGA, a total of 440 GC patients have data on genomic aberrations. Of these, 436 patients have mutation data, 438 patients have CNV data, and 436 patients have SV data. We investigated the genomic aberrations in cadherins that are significantly differentially expressed between STAD and normal samples. Our results demonstrated that the 16 selected cadherins were altered across 227 (~52% of the profiled) GC patients. Figure 3A shows the patients with mutations or CNVs/SVs in at least 1 of the 16 cadherins considered for analysis. Protocadherin 17 (PCDH17) was found to be the most commonly altered cadherin gene in GC (54 of 440 samples =12.3%) followed by Fat atypical cadherin 1 (FAT1) (53 of 440 samples =12%), whereas PCDHGA10 was the least commonly altered cadherin in GC patients (12 of 440 =2.7%). Furthermore, we analyzed the rate of single nucleotide variations (SNVs) and CNVs/structural variations (SVs) independently and found that SNVs are more common across all the cadherins analyzed except for CDH17 (Figure 3A). Interestingly, 10.5% of GC patients had SNVs, while 1.8% patients had CNVs/SVs in PCDH17. Similarly, 8% patients had SNPs in FAT1, whereas 4% had CNVs/SVs. Moreover, 3% GC patients had CNVs/SVs and 2.3% patients had SNPs in CDH17. As expected, missense mutations were more common across the GC patients for all genes. In addition to missense mutations, CDH1 harbored splice site mutations in a higher number of patients than the other cadherins. Furthermore, although 14 cadherins harbored both amplifications and deletions, CDH17 harbored only amplifications, and FAT1 harbored only deletions in GC patients.
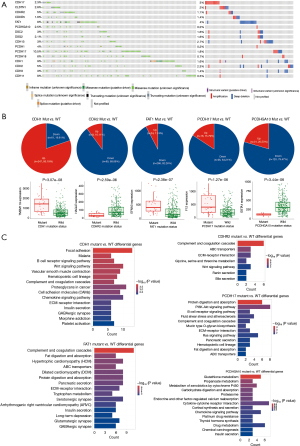
Next, we intended to explore whether mutations in these cadherins affect the expression of other genes. We used muTarget tool to assess the effect of mutant cadherins on the expression of other genes in GC. Our results revealed that mutations in 5 (CDH1, CDHR2, FAT1, PCDH17, and PCDHGA10) of the 16 genes affected the expression of other genes significantly (with log2FC >1 and FDR P<0.05) in GC (Figure 3B). The FAT1-mutant gene significantly affected the expression of most genes (n=310) followed by CDH1-mutant gene (n=308). Except in CDH1, mutation in all cadherins decreased the expression of the affected genes. Some 80% of the dysregulated genes in CDH1-mutant GC samples were upregulated, whereas less than 25% genes were upregulated in the other 4 cadherin-mutant samples. Figure 3B shows the most significantly dysregulated gene for each cadherin-mutant. The top 5 differentially expressed genes (DEGs) between cadherin-mutant and wildtype samples are shown in Table 2 (complete list of DEGs is provided in Supplementary file available at https://cdn.amegroups.cn/static/public/jgo-23-700-3.xlsx). To understand the effect of cadherin mutations at the molecular level, we performed pathway enrichment analysis of the downstream affected genes in cadherin-mutant versus cadherin-wildtype GC samples. The pathways that were significant with FDR P<0.1 were considered significant. Our results revealed that mutation of all five cadherins except PCDHGA10 mainly affected the ECM-receptor interaction pathway. Furthermore, mutation of all cadherins affected immune system-related pathways including ‘Complement and coagulation cascades’ being amongst the top 10 pathways (Figure 3C). Interestingly, mutation in FAT1 affected cardiac disease related pathways, such as cardiomyopathy. Adhesion- and inflammation-related genes are known to be involved in these cardiac pathways. Additionally, PCDHGA10-mutants were shown to mainly affect the metabolic pathways, including glutathione and propanoate metabolism. Thus, our analysis showed that mutations in cadherin genes commonly affect adhesion-related pathways by regulating the corresponding genes products. However, a few mutant cadherins were found to affect the cardiac and metabolic pathways and their significance in GC needs to be experimentally verified.
Table 2
| Gene | Log2 fold change (mutant/wildtype) | P value | P value (false discovery rate) |
|---|---|---|---|
| CDH1 mutant vs. wildtype | |||
| TMEM119 | 1.3 | 3.07E−08 | 4.11E−04 |
| OMD | 1.8 | 1.65E−07 | 6.29E−04 |
| PCOLCE | 1.0 | 3.06E−07 | 6.29E−04 |
| CYP1B1 | 1.7 | 3.34E−07 | 6.29E−04 |
| GAS1 | 1.2 | 4.87E−07 | 6.29E−04 |
| CDHR2 mutant vs. wildtype | |||
| HAGLR | −2.4 | 2.59E−06 | 5.22E−03 |
| DIPK1B | −1.1 | 7.94E−06 | 9.26E−03 |
| SYT8 | 1.5 | 8.30E−06 | 9.26E−03 |
| NR5A2 | −2.0 | 1.40E−05 | 1.11E−02 |
| ONECUT2 | −1.9 | 3.84E−05 | 1.41E−02 |
| FAT1 mutant vs. wildtype | |||
| EFNA3 | 1.3 | 2.38E−07 | 9.66E−04 |
| VTN | −3.0 | 4.30E−07 | 9.66E−04 |
| SERPINF2 | −2.7 | 4.38E−07 | 9.66E−04 |
| CREB3L3 | −2.1 | 5.78E−07 | 9.66E−04 |
| MMP24 | −1.5 | 6.49E−07 | 9.66E−04 |
| PCDHGA10 mutant vs. wildtype | |||
| GSTA4 | −2.0 | 3.44E−06 | 1.90E−02 |
| CAPN12 | −2.8 | 7.71E−06 | 1.90E−02 |
| IFITM1 | 1.5 | 8.03E−06 | 1.90E−02 |
| TNFSF9 | 2.4 | 8.92E−06 | 1.90E−02 |
| EIF5AL1 | 1.9 | 9.98E−06 | 1.90E−02 |
| PCDH17 mutant vs. wildtype | |||
| F12 | 1.1 | 1.27E−06 | 1.83E−03 |
| CARD11 | −1.5 | 1.79E−06 | 2.15E−03 |
| RGS5 | −1.0 | 3.60E−06 | 2.32E−03 |
| WASF3 | −1.1 | 5.08E−06 | 2.84E−03 |
| SLC7A2 | −2.5 | 8.98E−06 | 3.99E−03 |
Expression of cadherin genes is associated with survival of stomach cancer patients
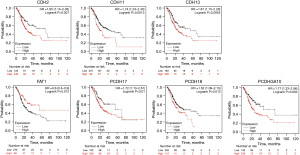
Further, we investigated whether these differentially expressed cadherins affect the OS of GC patients. Our results revealed that 7 (CDH2, CDH11, CDH13, FAT1, PCDH17, PCDH18, and PCDHGA10) of the 16 cadherins were significantly (P<0.05) associated with OS of STAD patients (Figure 4). Low expression at the mRNA level of all the cadherins except FAT1 correlated with higher survival rate. For instance, the median survival rate was 70 months when CDH2 expression is lower compared to 25 months when its expression is higher in GC patient cohorts (P=0.007). Similarly, patients with lower expression of PCDHGA10 survived 73 months compared to those with higher expression (median survival, 23 months; P=0.002). However, patients with lower expression of FAT1 survived 26 months as compared to those with higher expression, who survived 56 months (P=0.012). Thus, our analysis revealed that lower expression of most cadherins, yet higher expression of FAT1, is associated with better prognosis of GC patients. Additionally, expression levels of CDH11, FAT1, and PCDH17 were significantly associated with the survival rate of stomach cancer patients of different race or gender (Figures S1-S4).
Abundances of major immune cells are correlated with cadherin genes in stomach cancer
We aimed to estimate the immune cells infiltration in gastric cancer (GC) samples and then explore whether their abundance is correlated with the expression of selected cadherin genes in stomach cancer. Our analysis revealed that 11 of the 24 immune cells had significant differences (P<0.05) in their abundance between GC patients and normal controls (Figure 5A). Among these, B cells, dendritic cells (DCs), natural killer (NK) cells, CD4 T cells, and natural killer T (NKT) cells were considerably more abundant (median abundance >0.01) in both cancer and normal samples, whereas the remaining 6 cell types were less abundant in both conditions (median abundance <0.01). The abundance of B cells, DC, and NKT cells was higher in cancer samples, whereas that of NK cells and CD4-T cells was lower in cancer samples than in normal samples (Supplementary file available at https://cdn.amegroups.cn/static/public/jgo-23-700-4.xlsx).
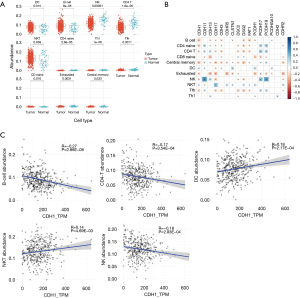
Further, we correlated the expression of 16 cadherin genes with the abundance of immune cell types that were significantly different (P<0.05) between cancer and normal stomach samples (Figure 5B). B cell abundance significantly correlated with 9 cadherin genes (all negative correlations), DC abundance correlated with 7 cadherin genes (6 positive and 1 negative), NKT cells correlated with 9 cadherin genes (8 positive and 1 negative), NK cells correlated with 9 genes (4 positive and 5 negative), and CD4 T cells correlated with 10 cadherins (4 positive and 6 negative). CDH1 and CDH17, each correlated significantly with all 5 highly abundant cell types (B cell, DC, NK, NKT, and CD4 T cell). Most of these were negative correlations. Interestingly, both CDH1 and CDH17 correlated positively with NKT cells. Figure 5C shows the correlation details for CDH1 with five highly abundant immune cell types (B cell, DC, NK cell, CD4 T cell, and NKT cell).
miRNA analysis revealed novel miRNA-cadherin interactions
We first identified miRNAs that are differentially expressed between GC and normal stomach samples based on TCGA miRNA-seq data. A total of 361 miRNAs (upregulated: 187; downregulated: 174) were significantly different between these two groups with absolute FC 1.5 (log2FC >0.58) and adjusted P<0.05 (Figure 6A). We then explored which of these upregulated miRNAs targeted the downregulated cadherins and vice versa in GC. Our results revealed that 9 upregulated miRNAs targeted the only downregulated cadherin, CDH2, whereas 79 downregulated miRNAs targeted the remaining 14 upregulated cadherins (CDHR2 was not targeted by any miRNA), resulting in a total of 135 miRNA-gene interactions. Further, using these interactions, we constructed a network to identify miRNAs that are targeting multiple cadherins. The network contained a total of 103 nodes and 135 edges. PCDH17 was targeted by the highest number of miRNAs (n=37) followed by CDH11 with 19 miRNAs (Figure 6B). Furthermore, 2 miRNAs (hsa-miR-23b-3p and hsa-miR-495-3p) targeted 4 cadherins each, whereas 5 miRNAs (has-miR-9-3p, has-let-7a-2-3p, has-miR-4775, has-miR-9-5p, has-miR-548ba) targeted 3 cadherins each (Table 3 and Supplementary file available at https://cdn.amegroups.cn/static/public/jgo-23-700-5.xlsx). The miRNA hsa-miR-23b-3p targeted CDH1, DSC2, PCDH17, and PCDH18, whereas hsa-miR-495-3p targeted CDH1, DSG2, CDH13, and PCDH18. Surprisingly, except for 1 miRNA-gene interaction (hsa-miR-548ba-PCDH17), none of the interactions have been experimentally verified according to the data obtained from miRTarBase, a database of experimentally verified miRNA-target interactions.
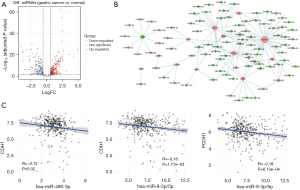
Table 3
| microRNA name | Log2 fold change | Cadherin symbol | Interaction in miRTarbase | |
|---|---|---|---|---|
| microRNA | Cadherin | |||
| hsa-miR-23b-3p | −0.98 | 1.60 | CDH1 | − |
| hsa-miR-9-3p | −2.23 | 1.60 | CDH1 | − |
| hsa-miR-495-3p | −0.65 | 1.60 | CDH1 | − |
| hsa-let-7a-2-3p | −0.89 | 4.91 | CDH17 | − |
| hsa-let-7a-2-3p | −0.89 | 1.95 | FAT1 | − |
| hsa-miR-4775 | −1.51 | 2.01 | DSC2 | − |
| hsa-miR-9-3p | −2.23 | 2.01 | DSC2 | − |
| hsa-miR-23b-3p | −0.98 | 2.01 | DSC2 | − |
| hsa-miR-495-3p | −0.65 | 2.65 | DSG2 | − |
| hsa-miR-495-3p | −0.65 | 1.72 | CDH13 | − |
| hsa-miR-9-5p | −1.49 | 1.52 | PCDH1 | − |
| hsa-miR-4775 | −1.51 | 1.19 | PCDH17 | − |
| hsa-miR-9-5p | −1.49 | 1.19 | PCDH17 | − |
| hsa-miR-23b-3p | −0.98 | 1.19 | PCDH17 | − |
| hsa-miR-548ba | −2.46 | 1.19 | PCDH17 | Yes |
| hsa-miR-9-3p | −2.23 | 1.19 | PCDH17 | − |
| hsa-miR-495-3p | −0.65 | 1.01 | PCDH18 | − |
| hsa-miR-23b-3p | −0.98 | 1.01 | PCDH18 | − |
| hsa-miR-548ba | −2.46 | 1.01 | PCDH18 | − |
| hsa-miR-4775 | −1.51 | 1.01 | PCDH18 | − |
| hsa-let-7a-2-3p | −0.89 | 2.28 | CDH11 | − |
| hsa-miR-9-5p | −1.49 | 2.28 | CDH11 | − |
| hsa-miR-548ba | −2.46 | 2.28 | CDH11 | − |
Log2 fold change correspond to difference in gastric cancer vs. normal. Negative fold change values indicate down-regulation, whereas positive values indicate up-regulation.
To further strengthen the regulatory role of the miRNAs by targeting the cadherin genes, we performed co-expression analysis between top miRNAs and cadherin-mRNA expression in GC by using normalized expression values. Of the 23 cadherin-miRNA pairs tested (Table 3), the correlation for many pairs was insignificant, but a few pairs positively correlated. We observed significant negative correlations for hsa-miR-495-3p-CDH1 (P=0.02), hsa-miR-9-3p-CDH1 (P=1.77E−03), and hsa-miR-9-5p-PCDH1 (P=6.13E−04) pairs (Figure 6C). The negative correlation was also observed for hsa-let-7a-2-3p-CDH11, hsa-let-7a-2-3p-CDH17, hsa-miR-23b-3p-CDH1, and hsa-miR-495-3p-DSG2 pairs, however the correlation values were statistically insignificant. Thus, our miRNA-cadherin interaction analysis indicated that cadherins are actively regulated by many miRNAs in GC and PCDH17 is the most commonly miRNA-regulated cadherin in GC. However, experimental validation of the miRNA-cadherin interactions is required to understand their functional significance.
Discussion
The cadherin family of genes are essential in maintaining the integrity and function of stomach tissues, including cell-cell adhesion, cell migration, and differentiation (16). Dysregulation of cadherin expression and function has been linked to various gastric diseases, including GC (22). Therefore, understanding the regulation of cadherin expression and function in the stomach is crucial for the development of targeted therapeutic strategies for gastric diseases. In this study, we investigated the expression, mutational status, functional importance, and prognostic potential of cadherin family genes in STAD.
In this study, we identified 16 cadherin genes that showed significant differential expression between GC and normal samples. CDH1 is an important player in epithelial-mesenchymal transition, a root cause of invasive and metastatic cancer cell spreading (49). The differential expression of CDH1 has been reported to vary depending upon the histological subtypes of GC (50). For instance, CDH1 is upregulated in intestinal type GC, whereas it is downregulated in the diffuse type of adenocarcinoma owing to mutation and epigenetic modifications (50). In the current study, we used TCGA samples for the differential expression analysis of cadherins. A total of 450 STAD tumor samples in TCGA had the histological classification available. Among these, 41% (186 samples) belong to intestinal type, whereas only 16% (71 samples) belong to diffuse type. A higher percentage of intestinal subtype in the TCGA-STAD sample pool could explain the upregulation of CDH1 in our study. Furthermore, we found that CDH17 was highly upregulated in STAD between tumor and normal samples. These findings are consistent with a previous study that has reported the upregulation of CDH17 in GC (27), and suggest that CDH17 may be a potential diagnostic or therapeutic target for this disease.
In addition, we identified several other cadherin genes that were highly upregulated in GC, including CDHR5, DSC2, DSG2, CDH3, and CDH11. These genes have been previously implicated in cancer cell invasion, migration, and metastasis. For example, up-regulation of CDHR5 expression promotes malignant phenotype of pancreatic ductal adenocarcinoma (51); DSC2 expression was significantly increased in prostate cancer cells (52); DSG2 has been identified as a biomarker that promotes tumor proliferation and metastasis and is positively correlated with poor prognosis in early-stage cervical cancer (53); CDH3 expression is upregulated in thyroid cancer tissues compared to the adjacent normal tissues, and siRNA-mediated downregulation of CDH3 has been shown to inhibit the growth, migration, and invasion of thyroid cancer cells (54). The upregulation of these genes could be indicative of increased tumor aggressiveness and poor patient outcomes. Future studies should explore the potential clinical utility of these genes as prognostic biomarkers or therapeutic targets in GC.
Interestingly, we found that CDH2, a classical cadherin gene that has been previously implicated in cancer cell invasion and metastasis (55,56) to be downregulated in GC patients compared to normal controls. This finding is somewhat unexpected given the established role of CDH2 in promoting cancer cell invasion and metastasis. It is possible that downregulation of CDH2 may be a compensatory mechanism to counteract the effects of other upregulated cadherin genes in GC. Alternatively, it may indicate a different role for CDH2 in the pathogenesis of GC, which could be clarified using siRNA-based gene silencing strategies.
The identification of genes that are correlated with differentially expressed cadherins and genes that are affected by mutated cadherins in GC allowed us to perform functional enrichment analysis and gain insights into the functional importance of these cadherins in GC. Our analysis revealed that the correlated genes for most of the cadherins were involved in focal adhesion, ECM-receptor interaction, or junction-related pathways, all of which are related to cell adhesion. The focal adhesion and ECM-receptor interaction pathways play an important role in cell migration, proliferation, survival, and differentiation of cancer cells (57,58), and their dysregulation has been implicated in many pathological conditions, including cancer (59,60). The junction-related pathways are important for coordinating cell behavior, maintaining tissue integrity, and regulating the transport of ions and small molecules across epithelial and endothelial cell layers (61-64). Dysfunction of these junctions can have significant impacts on tissue and organ function and may contribute to the development of various diseases, including inflammation, cancer progression, and tumor metastasis (65-67).
Additionally, our functional enrichment analysis showed that the downstream genes for all mutated cadherins in GC were involved in the complement and coagulation cascades pathway. The complement system is a group of proteins that act to defend against invading pathogens by marking them for destruction and recruiting other immune cells to the site of infection (68-70). The coagulation system, on the other hand, is responsible for forming blood clots to stop bleeding after injury (71-74). Both systems are tightly regulated to prevent excessive immune activation or clot formation, which can lead to harmful effects. Furthermore, complement proteins can bind to ECM components such as laminin, fibronectin, and collagen, playing a role in immune surveillance and defense against invading pathogens (75,76). Coagulation factors, such as fibrinogen also interact with ECM and promote clotting (77,78). Dysregulation of these pathways can have serious consequences, and result in autoimmune diseases or thrombosis (79-83). Therefore, understanding the regulation of these pathways is key to the study of molecular mechanisms of cadherins during development of GC.
Finally, our immune cell infiltration analysis showed that 11 immune cell types have significantly different abundance between GC patients and normal controls, with B cells, DCs, and NKT cells being more abundant in cancer samples, whereas NK cells and CD4 T cells were less abundant. Correlating the expression of 16 cadherin genes with the abundance of immune cell types revealed that B cell abundance was negatively correlated with all 9 cadherin genes, whereas DC and NKT cell abundance was positively correlated with a majority of the cadherin genes. These findings further support interaction between immune cell types and the expression of cadherin genes in GC.
We found that about 45% of DEGs were significantly associated with the survival of STAD patients. Lower mRNA levels of most cadherins were correlated with higher survival rates. Reduced cadherin expression can contribute to weakened cell-cell adhesion, leading to decreased tumor cell migration, invasion, and metastasis. This could result in better containment of the tumor and slower disease progression.
Moreover, low cadherin expression may also reflect a less dedifferentiated state of cancer cells. High levels of cadherins are associated with epithelial-to-mesenchymal transition (EMT), a process whereby epithelial cells acquire more migratory and invasive properties (26,84). In contrast, low cadherin expression suggests a more epithelial phenotype, which is typically associated with a better prognosis in GC.
It is important to note that the relationship between cadherin expression and survival rates in GC is complex and can vary depending on the specific cadherin and the molecular context of the tumor. Further experimental validations are needed to fully understand the underlying mechanisms and potential therapeutic implications of cadherin expression in GC patients.
Abnormal expression of miRNAs has been observed in GC tissues compared to normal stomach tissues. These dysregulated miRNAs can act as oncogenes or tumor suppressors, influencing various cellular processes involved in cancer development, progression, and metastasis (85-89). Part of cadherin gene expression change in GC could result through miRNAs. We found that a total of 361 miRNAs exhibited significant differences between GC and normal samples. The miRNA-mRNA network analysis revealed that 9 upregulated miRNAs targeted CDH2, whereas 79 downregulated miRNAs targeted the upregulated cadherins (excluding CDHR2). These findings suggest that multiple miRNAs actively regulate cadherins in GC. However, further validations are warranted to clarify the functional significance of these miRNA-cadherin interactions in GC.
Conclusions
In conclusion, our study provides valuable insights into the alterations of cadherin genes in GC and their downstream effects on gene expression and pathways. However, further experimental validations are needed to elucidate the exact mechanisms by which these cadherins contribute to the development and progression of GC and their potential clinical implications.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-700/rc
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-700/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-700/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Dicken BJ, Bigam DL, Cass C, et al. Gastric adenocarcinoma: review and considerations for future directions. Ann Surg 2005;241:27-39. [Crossref] [PubMed]
- Plummer M, Franceschi S, Vignat J, et al. Global burden of gastric cancer attributable to Helicobacter pylori. Int J Cancer 2015;136:487-90. [Crossref] [PubMed]
- Dong J, Thrift AP. Alcohol, smoking and risk of oesophago-gastric cancer. Best Pract Res Clin Gastroenterol 2017;31:509-17. [Crossref] [PubMed]
- Rota M, Pelucchi C, Bertuccio P, et al. Alcohol consumption and gastric cancer risk-A pooled analysis within the StoP project consortium. Int J Cancer 2017;141:1950-62. [Crossref] [PubMed]
- Nagtegaal ID, Odze RD, Klimstra D, et al. The 2019 WHO classification of tumours of the digestive system. Histopathology 2020;76:182-8. [Crossref] [PubMed]
- Rawla P, Barsouk A. Epidemiology of gastric cancer: global trends, risk factors and prevention. Prz Gastroenterol 2019;14:26-38. [Crossref] [PubMed]
- Song Z, Wu Y, Yang J, et al. Progress in the treatment of advanced gastric cancer. Tumour Biol 2017;39:1010428317714626. [Crossref] [PubMed]
- Thrift AP, El-Serag HB. Burden of Gastric Cancer. Clin Gastroenterol Hepatol 2020;18:534-42. [Crossref] [PubMed]
- Pimentel-Nunes P, Dinis-Ribeiro M, Ponchon T, et al. Endoscopic submucosal dissection: European Society of Gastrointestinal Endoscopy (ESGE) Guideline. Endoscopy 2015;47:829-54. [Crossref] [PubMed]
- Smyth EC, Verheij M, Allum W, et al. Gastric cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2016;27:v38-49. [Crossref] [PubMed]
- Zhang Z, Xie T, Zhang X, et al. Immune checkpoint inhibitors for treatment of advanced gastric or gastroesophageal junction cancer: Current evidence and future perspectives. Chin J Cancer Res 2020;32:287-302. [Crossref] [PubMed]
- van Roy F, Berx G. The cell-cell adhesion molecule E-cadherin. Cell Mol Life Sci 2008;65:3756-88. [Crossref] [PubMed]
- van Roy F. Beyond E-cadherin: roles of other cadherin superfamily members in cancer. Nat Rev Cancer 2014;14:121-34. [Crossref] [PubMed]
- Yagi T, Takeichi M. Cadherin superfamily genes: functions, genomic organization, and neurologic diversity. Genes Dev 2000;14:1169-80. [Crossref] [PubMed]
- Halbleib JM, Nelson WJ. Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev 2006;20:3199-214. [Crossref] [PubMed]
- Katz BZ, Levenberg S, Yamada KM, et al. Modulation of cell-cell adherens junctions by surface clustering of the N-cadherin cytoplasmic tail. Exp Cell Res 1998;243:415-24. [Crossref] [PubMed]
- Stemmler MP. Cadherins in development and cancer. Mol Biosyst 2008;4:835-50. [Crossref] [PubMed]
- Berx G, van Roy F. Involvement of members of the cadherin superfamily in cancer. Cold Spring Harb Perspect Biol 2009;1:a003129. [Crossref] [PubMed]
- Katoh M. Function and cancer genomics of FAT family genes Int J Oncol 2012;41:1913-8. (review). [Crossref] [PubMed]
- Yakirevich E, Resnick MB. Pathology of gastric cancer and its precursor lesions. Gastroenterol Clin North Am 2013;42:261-84. [Crossref] [PubMed]
- Carneiro F. Familial and hereditary gastric cancer, an overview. Best Pract Res Clin Gastroenterol 2022;58-59:101800. [Crossref] [PubMed]
- David JM, Rajasekaran AK. Dishonorable discharge: the oncogenic roles of cleaved E-cadherin fragments. Cancer Res 2012;72:2917-23. [Crossref] [PubMed]
- Semb H, Christofori G. The tumor-suppressor function of E-cadherin. Am J Hum Genet 1998;63:1588-93. [Crossref] [PubMed]
- Gottardi CJ, Wong E, Gumbiner BM. E-cadherin suppresses cellular transformation by inhibiting beta-catenin signaling in an adhesion-independent manner. J Cell Biol 2001;153:1049-60. [Crossref] [PubMed]
- Loh CY, Chai JY, Tang TF, et al. The E-Cadherin and N-Cadherin Switch in Epithelial-to-Mesenchymal Transition: Signaling, Therapeutic Implications, and Challenges. Cells 2019;8:1118. [Crossref] [PubMed]
- Long ZW, Zhou ML, Fu JW, et al. Association between cadherin-17 expression and pathological characteristics of gastric cancer: a meta-analysis. World J Gastroenterol 2015;21:3694-705. [Crossref] [PubMed]
- Hansford S, Kaurah P, Li-Chang H, et al. Hereditary Diffuse Gastric Cancer Syndrome: CDH1 Mutations and Beyond. JAMA Oncol 2015;1:23-32. [Crossref] [PubMed]
- Donner I, Kiviluoto T, Ristimäki A, et al. Exome sequencing reveals three novel candidate predisposition genes for diffuse gastric cancer. Fam Cancer 2015;14:241-6. [Crossref] [PubMed]
- Lobo S, Benusiglio PR, Coulet F, et al. Cancer predisposition and germline CTNNA1 variants. Eur J Med Genet 2021;64:104316. [Crossref] [PubMed]
- Im S, Cho YK, Kang D, et al. Combined high NEDD9 expression and E-cadherin loss correlate with poor clinical outcome in gastric cancer. J Gastroenterol Hepatol 2022;37:2255-63. [Crossref] [PubMed]
- Fang WK, Liao LD, Li LY, et al. Down-regulated desmocollin-2 promotes cell aggressiveness through redistributing adherens junctions and activating beta-catenin signalling in oesophageal squamous cell carcinoma. J Pathol 2013;231:257-70. [Crossref] [PubMed]
- Tang Z, Kang B, Li C, et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019;47:W556-60. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013;45:1113-20. [Crossref] [PubMed]
- Carithers LJ, Moore HM. The Genotype-Tissue Expression (GTEx) Project. Biopreserv Biobank 2015;13:307-8. [Crossref] [PubMed]
- Liao Y, Wang J, Jaehnig EJ, et al. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res 2019;47:W199-205. [Crossref] [PubMed]
- Li J, Miao B, Wang S, et al. Hiplot: a comprehensive and easy-to-use web service for boosting publication-ready biomedical data visualization. Brief Bioinform 2022;23:bbac261. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Nagy Á, Győrffy B. muTarget: A platform linking gene expression changes and mutation status in solid tumors. Int J Cancer 2021;148:502-11. [Crossref] [PubMed]
- Nagy Á, Munkácsy G, Győrffy B. Pancancer survival analysis of cancer hallmark genes. Sci Rep 2021;11:6047. [Crossref] [PubMed]
- Lánczky A, Győrffy B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J Med Internet Res 2021;23:e27633. [Crossref] [PubMed]
- Chandrashekar DS, Karthikeyan SK, Korla PK, et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022;25:18-27. [Crossref] [PubMed]
- Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res 2020;48:D127-31. [Crossref] [PubMed]
- Huang HY, Lin YC, Cui S, et al. miRTarBase update 2022: an informative resource for experimentally validated miRNA-target interactions. Nucleic Acids Res 2022;50:D222-30. [Crossref] [PubMed]
- Xu F, Wang Y, Ling Y, et al. dbDEMC 3.0: Functional Exploration of Differentially Expressed miRNAs in Cancers of Human and Model Organisms. Genomics Proteomics Bioinformatics 2022;20:446-54. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Miao YR, Zhang Q, Lei Q, et al. ImmuCellAI: A Unique Method for Comprehensive T-Cell Subsets Abundance Prediction and its Application in Cancer Immunotherapy. Adv Sci (Weinh) 2020;7:1902880. [Crossref] [PubMed]
- Bure IV, Nemtsova MV, Zaletaev DV. Roles of E-cadherin and Noncoding RNAs in the Epithelial-mesenchymal Transition and Progression in Gastric Cancer. Int J Mol Sci 2019;20:2870. [Crossref] [PubMed]
- Tanabe S, Aoyagi K, Yokozaki H, et al. Gene expression signatures for identifying diffuse-type gastric cancer associated with epithelial-mesenchymal transition. Int J Oncol 2014;44:1955-70. [Crossref] [PubMed]
- Gao J, Wang M, Li T, et al. Up-regulation of CDHR5 expression promotes malignant phenotype of pancreatic ductal adenocarcinoma. J Cell Mol Med 2020;24:12726-35. [Crossref] [PubMed]
- Jiang F, Wu P. Regulating DSC2 Expression Affects the Proliferation and Apoptosis of Prostate Cancer Cells. Cancer Manag Res 2020;12:11453-62. [Crossref] [PubMed]
- Qin S, Liao Y, Du Q, et al. DSG2 expression is correlated with poor prognosis and promotes early-stage cervical cancer. Cancer Cell Int 2020;20:206. [Crossref] [PubMed]
- Zhou Y, Chi Y, Bhandari A, et al. Downregulated CDH3 decreases proliferation, migration, and invasion in thyroid cancer. Am J Transl Res 2020;12:3057-67. [PubMed]
- Ma T, Zhao Y, Wei K, et al. MicroRNA-124 Functions as a Tumor Suppressor by Regulating CDH2 and Epithelial-Mesenchymal Transition in Non-Small Cell Lung Cancer. Cell Physiol Biochem 2016;38:1563-74. [Crossref] [PubMed]
- Liu T, Xia R, Li C, et al. mRNA expression level of CDH2, LEP, POSTN, TIMP1 and VEGFC modulates 5-fluorouracil resistance in colon cancer cells. Exp Ther Med 2021;22:1023. [Crossref] [PubMed]
- Wu C. Focal adhesion: a focal point in current cell biology and molecular medicine. Cell Adh Migr 2007;1:13-8. [Crossref] [PubMed]
- Walma DAC, Yamada KM. The extracellular matrix in development. Development 2020;147:dev175596. [Crossref] [PubMed]
- Bao Y, Wang L, Shi L, et al. Transcriptome profiling revealed multiple genes and ECM-receptor interaction pathways that may be associated with breast cancer. Cell Mol Biol Lett 2019;24:38. [Crossref] [PubMed]
- Zhang HJ, Tao J, Sheng L, et al. Twist2 promotes kidney cancer cell proliferation and invasion by regulating ITGA6 and CD44 expression in the ECM-receptor interaction pathway. Onco Targets Ther 2016;9:1801-12. [PubMed]
- Goodenough DA, Paul DL. Gap junctions. Cold Spring Harb Perspect Biol 2009;1:a002576. [Crossref] [PubMed]
- Hartsock A, Nelson WJ. Adherens and tight junctions: structure, function and connections to the actin cytoskeleton. Biochim Biophys Acta 2008;1778:660-9. [Crossref] [PubMed]
- Harris TJ, Tepass U. Adherens junctions: from molecules to morphogenesis. Nat Rev Mol Cell Biol 2010;11:502-14. [Crossref] [PubMed]
- Anderson JM, Van Itallie CM. Physiology and function of the tight junction. Cold Spring Harb Perspect Biol 2009;1:a002584. [Crossref] [PubMed]
- Bonacquisti EE, Nguyen J. Connexin 43 (Cx43) in cancer: Implications for therapeutic approaches via gap junctions. Cancer Lett 2019;442:439-44. [Crossref] [PubMed]
- Chen Q, Boire A, Jin X, et al. Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature 2016;533:493-8. [Crossref] [PubMed]
- Roehlen N, Roca Suarez AA, El Saghire H, et al. Tight Junction Proteins and the Biology of Hepatobiliary Disease. Int J Mol Sci 2020;21:825. [Crossref] [PubMed]
- Müller-Eberhard HJ. Molecular organization and function of the complement system. Annu Rev Biochem 1988;57:321-47. [Crossref] [PubMed]
- Morgan BP, Walters D, Serna M, et al. Terminal complexes of the complement system: new structural insights and their relevance to function. Immunol Rev 2016;274:141-51. [Crossref] [PubMed]
- Pouw RB, Ricklin D. Tipping the balance: intricate roles of the complement system in disease and therapy. Semin Immunopathol 2021;43:757-71. [Crossref] [PubMed]
- Berg DE. Components and defects of the coagulation system. Nurse Pract Forum 1992;3:62-71. [PubMed]
- Chee YL. Coagulation. J R Coll Physicians Edinb 2014;44:42-5. [Crossref] [PubMed]
- Matsumoto T, Nogami K, Shima M. Coagulation function and mechanisms in various clinical phenotypes of patients with acquired factor V inhibitors. J Thromb Haemost 2014;12:1503-12. [Crossref] [PubMed]
- Brake MA, Ivanciu L, Maroney SA, et al. Assessing Blood Clotting and Coagulation Factors in Mice. Curr Protoc Mouse Biol 2019;9:e61. [Crossref] [PubMed]
- Silawal S, Triebel J, Bertsch T, et al. Osteoarthritis and the Complement Cascade. Clin Med Insights Arthritis Musculoskelet Disord 2018;11:1179544117751430. [Crossref] [PubMed]
- Kalchishkova N, Fürst CM, Heinegård D, et al. NC4 Domain of cartilage-specific collagen IX inhibits complement directly due to attenuation of membrane attack formation and indirectly through binding and enhancing activity of complement inhibitors C4B-binding protein and factor H. J Biol Chem 2011;286:27915-26. [Crossref] [PubMed]
- Oh H, Park HE, Song MS, et al. The Therapeutic Potential of Anticoagulation in Organ Fibrosis. Front Med (Lausanne) 2022;9:866746. [Crossref] [PubMed]
- Posner MG. Multimerin-1 and cancer: a review. Biosci Rep 2022;42:BSR20211248. [Crossref] [PubMed]
- Morgan BP. The role of complement in neurological and neuropsychiatric diseases. Expert Rev Clin Immunol 2015;11:1109-19. [Crossref] [PubMed]
- Vignesh P, Rawat A, Sharma M, et al. Complement in autoimmune diseases. Clin Chim Acta 2017;465:123-30. [Crossref] [PubMed]
- Menegatti M, Palla R. Clinical and laboratory diagnosis of rare coagulation disorders (RCDs). Thromb Res 2020;196:603-8. [Crossref] [PubMed]
- Pekna M, Pekny M. The Complement System: A Powerful Modulator and Effector of Astrocyte Function in the Healthy and Diseased Central Nervous System. Cells 2021;10:1812. [Crossref] [PubMed]
- Bohania N, Agrawal A, Prakash A, et al. Coagulation Profile and its Correlation with Severity of Liver Dysfunction and Gastrointestinal Bleed in Alcoholic Liver Disease Patients. J Assoc Physicians India 2021;69:11-2. [PubMed]
- Wheelock MJ, Shintani Y, Maeda M, et al. Cadherin switching. J Cell Sci 2008;121:727-35. [Crossref] [PubMed]
- Ruan K, Fang X, Ouyang G. MicroRNAs: novel regulators in the hallmarks of human cancer. Cancer Lett 2009;285:116-26. [Crossref] [PubMed]
- Chun-Zhi Z, Lei H, An-Ling Z, et al. MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer 2010;10:367. [Crossref] [PubMed]
- Tsukamoto Y, Nakada C, Noguchi T, et al. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res 2010;70:2339-49. [Crossref] [PubMed]
- Feng L, Xie Y, Zhang H, et al. miR-107 targets cyclin-dependent kinase 6 expression, induces cell cycle G1 arrest and inhibits invasion in gastric cancer cells. Med Oncol 2012;29:856-63. [Crossref] [PubMed]
- Ding L, Xu Y, Zhang W, et al. MiR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res 2010;20:784-93. [Crossref] [PubMed]


