
Lymph node metastasis-related lncRNA GAS6-AS1 facilitates the progression of esophageal squamous cell carcinoma
Introduction
Esophageal cancer is a common malignant tumor, ranking seventh in incidence and sixth in mortality (1). Esophageal squamous cell carcinoma (ESCC) is the most common pathological type, especially in high-incidence areas of eastern Asia and in eastern and southern Africa (2), with over 50% of esophageal cancer cases occurring in China (3). Prognosis analysis of ESCC has shown that the 5-year overall survival (OS) was about 20% in the past decade in China (4) and the USA (5). Multiple factors affect the prognosis of ESCC, and the most important prognostic factors include tumor stage and histology (6). Recurrence and metastasis are the most important causes of poor prognosis in ESCC, with lymph node metastasis being a key indicator. According to research, the 5-year survival rate of ESCC after lymph node metastasis has decreased from 43% to 23% (7). The proportion of lymph node metastasis in ESCC is extremely high, especially when the primary tumor stage is relatively advanced (8). It has been reported that when the primary tumor invasion depth reaches above the adventitia layer (T3), lymph node metastasis exceeds 82.1% (9). Research on biomarkers that can effectively predict lymph node metastasis is an important means of improving the treatment of ESCC, which can effectively improve the prognosis of patients (10). Detecting cell free DNA levels in esophageal cancer is a potentially valuable tool for early detection and evaluation of the prognosis of esophageal cancer patients (11). With the advent of the next generation of sequencing technology, transcriptome sequencing (RNA-seq) has become a powerful tool (12) that has the unique ability to comprehensively study gene transcriptome data recognition. At present, it has been used for biomarker detection and personalized treatment of tumor patients. RNA-seq has been used to detect the expression level of long non-coding RNA (lncRNA) and study their related mechanisms of action. LncRNAs are a large class of non-protein-coding transcripts with a length greater than 200 nucleotides. Current research has shown that they are related to epigenetic regulation (13) and regulatory proteins (14) in cells, especially in cancer cells. For example, lncRNA urothelial carcinoma-associated 1 (UCA1), are oncogenes involved in regulating esophageal cancer progression (15). Previous studies have shown that lncRNAs can promote tumor lymph node metastasis by regulating the activity of various oncogenes, for example, a study has shown VESTAR, a lncRNA carried in SCNA, regulates the stability of VEGFC messenger RNA (mRNA), and promotes lymphatic vessel and lymphatic ESCC metastasis (16). In addition, a study has suggested that lncRNA can be used to predict lymph node metastasis in tumors, such as ELNAT1 being used to predict lymph node metastasis in breast tumors (17).
The relationship between lncRNA and lymph node metastasis in ESCC is still unclear. A previous study has shown that the expression of lncRNA BANCR gene significantly increased in tumor tissue, and overexpression of lncRNA BANCR was positively correlated with lymph node metastasis (18). We obtained some important lncRNAs through RNA-seq data analysis of T3 stage cancer tissues with and without lymph node metastasis, including LINC02381, lncRNA GAS6-AS1. To our knowledge, there is no similar tissue sequencing data. GAS6 antisense RNA 1 (GAS6-AS1) is a 902 bp lncRNA composed of 5 exons and transcribes GAS6-AS1 in antisense direction (19). To date, research has shown that it is closely related to the growth and metastasis of cancer. A study has shown that overexpression of GAS6-AS1 can inhibit tumor progression of lung adenocarcinoma (LUAD) both in vivo and in vitro, which is also associated with adverse clinical prognosis (20). This functional phenomenon has similar research results in renal papillary cell carcinoma, whereby high expression is associated with better survival in renal cell carcinoma (21). On the contrary, a study has shown that the high expression of GAS6-AS1 is closely related to the tumor staging of gastric cancer, and in vitro and in vivo experiments have shown that it can promote tumor growth, metastasis, and cell cycle changes (19). This is similar to the results of research on acute myeloid leukemia (AML), in which abnormal function leads to a more aggressive leukemia phenotype and poorer survival outcomes (22).
At present, the specific mechanism of GAS6-AS1 affecting tumor growth and metastasis remains unclear. Most studies have shown that the GAS6-AS1 is mainly involved in the pathogenesis of human cancer through GAS6-dependent or GAS6-independent mechanisms (19,23). GAS6 belongs to the vitamin K-dependent (VKD) family, which is located on chromosome 13q34, has 15 exons, and mainly regulates proliferation, migration, and apoptosis of various types of cancer (24). Tumor-related mechanism study has shown that GAS6-AS1 may promote the proliferation, migration, and invasion of gastric cancer cells in vitro and in vivo by up regulating its homeotic gene GAS6 (19), which mainly drives the activation of the AXL signaling pathway. In addition, research in clear cell renal cell carcinoma has shown that GAS6-AS1 regulates cell proliferation, invasion, and glycolysis by regulating the AMPK/mTOR signaling pathway (25). Research on leukemia has shown that the targeted therapy of the GAS6-AS1/YBX1/MYC axis inhibits AML cell proliferation and disease progression (22). One more study has shown that this lncRNA can serve as a sponge for miR-24-3p (26) to regulate GIMAP6. Therefore, the interaction between GAS6-AS1 and microRNA (miRNA) is the most likely pathway for GAS6-AS1 to participate in tumor development.
However, there is currently no research on the relationship between GAS6-AS1 and ESCC, especially its relationship with lymph node metastasis, and its potential clinical application value. This study firstly reported the comprehensive sequencing of transcriptome in ESCC tissues with or without lymph node metastasis in T3 phase by using RNA-seq. In this study, we revealed through RNA-seq screening and identification that lncRNA GAS6-AS1 is closely related to lymph node metastasis in ESCC, and demonstrated its potential as a new biomarker for predicting lymph node metastasis in ESCC. We present this article in accordance with the MDAR and ARRIVE reporting checklists (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-798/rc).
Methods
Clinical samples
A total of 187 ESCC tissues and 35 normal esophageal epithelial tissue were collected from ESCC patients undergoing surgery for esophageal carcinoma at Gaozhou People’s Hospital between January 2016 and June 2020. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study involving human experiments was approved by the Medical Ethics Committee of the Gaozhou People’s Hospital (No. GYLLYJ-2022086). Written informed consent was provided by all patients.
RNA sequencing
Firstly, total RNA extracts of ESCC and paired peritumoral normal tissues (n=10) were used for RNA-seq to identify the differentially expressed RNAs (including mRNAs and lncRNAs). Library preparation and RNA-seq was performed on an Illumina platform by Cheerland Biotechnology (Shenzhen, China). Differential expression analysis between the two groups was performed with limma R package. A linear model was used to fit and the Empirical Bayes test was used to obtain the P value for the test. The cutoff value of differentially expressed RNAs was set as |log2[fold change (FC)]| >0.585 and P<0.05.
RNA extraction and real-time quantitative polymerase chain reaction
Total RNA was extracted using a Cell/Tissue Total RNA Kit (19221ES50; Yeasen, Shanghai, China) from tissue or cultured cell lines, which was then applied to reverse transcription using a complementary DNA (cDNA) Synthesis Kit (11141ES60; Yeasen). Quantitative real-time polymerase chain reaction (qRT-PCR) was conducted using a Hieff UNICON® Universal Blue qPCR SYBR Green Master Mix (11184ES08; Yeasen). Expression data were standardized to the reference gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH) in order to control the variability in expression levels. The average for each gene and sample was calculated and the experiments were independently repeated three times (the primers used are listed in Table S1).
Cell lines
Human ESCC cell lines TE-1/KYSE410 were maintained in Roswell Park Memorial Institute (RPMI) 1640 medium (72400047; Gibco, Waltham, MA, USA) containing 10% fetal bovine serum (FBS; A3161002C; Gibco), human esophageal epithelial cells HET-1A was also maintained in RPMI 1640 medium containing 10% FBS. All wares were cultured at 37 ℃ and 5% CO2. TE-1/KYSE410 and HET-1A cells were authenticated by karyotype, morphology, and PCR analysis.
Construction of stable cell lines
To establish GAS6-AS1 knockdown stable cell lines, the human GAS6-AS1 short hairpin RNA (shRNA) plasmids were purchased from Guangzhou IGE Biotechnology Ltd. (Guangzhou, China) (sequences shown in Table S2). The lentiviral particles were used to transfect the KYSE410 cells by using Lenti-PacTM HIV lentiviral particles packaging Kit (LT003, GeneCopoeia, Guangzhou, China). The efficiency of the knockdown of GAS6-AS1 was determined by qRT-PCR.
Cell counting kit‑8 (CCK-8) cell viability assay
TE-1, KYSE410 cells were gathered 48 hours after transfection, processed with trypsin, and seeded into 96-well plates at a thickness of 1×104/well. In the wake of refined for 24, 48, and 72 hours, 10 µL of CCK-8 (KGA317s-1000; Keygen, Nanjing, China) stock arrangement was added to each well, at that point cells were incubated at 37 ℃ with 5% CO2 for 4 hours. Optical density (OD) values were then detected at a wavelength of 450 nm.
Colony formation assay
To perform colony formation assay, 500 cells/2 mL were inoculated into a 6-well plate and the culture medium was changed weekly. After 2 weeks, these cells were washed with phosphate buffered saline (PBS), fixed with paraformaldehyde for 20 minutes, and stained with 2% crystal violet for 30 minutes to calculate the bacterial count. These experiments were independently repeated three times.
Wound healing migration assay
For the measurement of wound healing migration, the fused monolayer cells were injured at the tip of a p20 pipette. These images were taken after PBS washing for three times (time 0 hours). And then cells were cultured in standard medium, and the wound healing rates after 24, and 48 hours were recorded. A total of three separate fields were captured for each board (one representative image is shown). All experiments were independently repeated three times.
Cell apoptosis and cell cycle assay
Cell apoptosis and cell cycle were respectively detected by the Annexin V-FITC/PI Double Staining Cell Apoptosis Detection Kit (KGA108; Keygen) and a Cell Cycle Detection Kit (KGA512; Keygen), in accordance to the manufacturer’s manual. Briefly, the cells were grown to 70% confluency and after 48 hours, the cells were collected and processed for analysis. The cell samples were then analyzed using a flow cytometer ACEA NovoCyte equipped with the FlowJo software [version 10; Becton, Dickinson, and Co. (BD), Franklin Lakes, NJ, USA].
Xenograft tumor model
BALB/c mice (3-week-old, male) were purchased from the Animal Experiment Center of Southern Medical University (Guangzhou, China) and maintained under standard conditions. Tumor cells of KYSE410 negative control (NC) and sh#GAS6-AS1 groups (2×106 cells/tumor in were suspended in 100 µL RPMI-1640) were injected into eight mice left and right axillary areas. The mice were observed every 2 days, and the experiment was finished at 18 days after tumor cell inoculation. Tumor volume was determined using the standard formula: L × W2/2, L: longest diameters, W: shortest diameters. A protocol was prepared before the study without registration. The animal experiments were approved by the Ethics Committee of the Gaozhou People’s Hospital (No. GYLLYJ-2022109), in compliance with national guidelines for the care and use of animals.
Statistical analyses
Statistical analyses were performed by GraphPad Prism version 9.0 (GraphPad Software, San Diego, CA, USA) and SPSS 26.0 (IBM Corp., Armonk, NY, USA). Quantitative data were presented as the mean ± standard deviation (SD). A χ2 test was performed to compare nonparametric variables. A 2-tailed unpaired/paired Student t-test was used for the comparison of parametric variables. The Kaplan-Meier (KM) analysis and log-rank test method were used to assess the OS and disease-free survival (DFS) of patients, respectively. Univariate and multivariate logistic regression analyses were performed, and the factors with P values less than 0.05 in univariate analysis were included in a multivariate logistic regression analysis. Multivariate analysis was used to identify independent risk factors for predicting lymph node metastasis. We used receiver operating characteristic (ROC) curves to evaluate predictive value, the area under the receiver operating characteristic curve (AUC) was applied to determine the discriminative values of GAS6-AS1 expression for lymph node metastasis.
Results
Differential profiling data based on RNA-seq related to lymph node metastasis
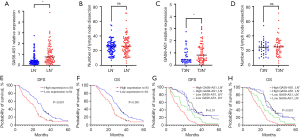
In order to find the transcriptome genes related to the development and progression of ESCC, especially those related to lymph node metastasis, we selected 10 patients with ESCC who underwent surgery. Their primary tumor stage was T3, which invaded the esophageal fibrous membrane. Among them, five had lymph node metastasis, and the other five had the same primary tumor stage without lymph node metastasis. The pathological type of all 10 cases was ESCC, and the degree of differentiation was medium to high, with no difference between gender and age (Table 1). Notably, they all underwent minimally invasive McKeown surgical treatment within the same thoracic surgery group of Gaozhou People’s Hospital, and there was no difference in the number of lymph nodes dissection between the two groups (Figure 1A). We performed RNA-seq on 10 pairs of ESCC tissues and adjacent normal tissues, and created a transcripts per million (TPM) distribution violin diagram of these 20 tissues (Figure 1B). At the 1.5-fold cutoff, a majority of 652 significantly changed genes were identified, and the differential expression of hierarchical clustering heat map is shown in Figure 1C. In order to further reduce candidate genes, we further analyzed the RNA-seq data of T3N+ group (primary tumor stage T3 with lymph node metastasis) and T3N− group (primary tumor stage T3 without lymph node metastasis), identified 159 significantly changed genes, and made a hierarchical clustering heat map of differential expression (Figure 1D). Through the analysis of two sets of data, we decided to display the intersection of the two sets of data through Venn diagram (Figure 1E) and identified 26 significantly altered RNAs, including lncRNA GAS6-AS1. Other genes related to esophageal cancer, such as LINC02381, and mRNA SIM2 has been confirmed to be associated with the growth and metastasis of esophageal cancer (27). Regarding the study of biological functions, we conducted Gene Ontology (GO) analysis on 10 pairs of differentially expressed genes (DEGs) between ESCC tissue and adjacent normal tissues. Important GO analysis aggregates showed biological processes and molecular functions as shown in Figure 1F. Most biological processes and molecular functions were associated with tumorigenesis, such as anti-apoptosis, cell migration, DNA replication and repair, and cell cycle regulation (28). For the T3N+ group and T3N− group, RNA-seq data analysis focused on tumor metastasis, such as cell migration, DNA replication and repair (Figure 1G). The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis tool was used for gene pathway analysis, which suggested that differential genes between cancer and normal tissues are associated with tumor growth and metastasis, such as the cAMP signaling pathway (29) and the cGMP PKG signaling pathway (30) (Figure 1H). RNA-seq data analysis of T3N+ and T3N− in cancer tissue suggested signal pathways related to tumor metastasis, such as the MAPK signaling pathway (31) and the N-Glycan biosynthesis pathway (32) (Figure 1I). The above results indicate that the intersection of sequencing data is related to tumor growth and metastasis.
Table 1
| Clinical parameter | Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | Patient 7 | Patient 8 | Patient 9 | Patient 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Gender | Male | Male | Female | Female | Male | Male | Male | Male | Female | Female |
| Age (years) | 79 | 77 | 57 | 67 | 66 | 67 | 83 | 78 | 49 | 71 |
| Pathological types | ESCC | ESCC | ESCC | ESCC | ESCC | ESCC | ESCC | ESCC | ESCC | ESCC |
| Differentiation grade | Moderate | Moderate | Moderate | Moderate | Moderate | Moderate | Moderate | Moderate | Moderate | Well |
| Primary tumor invasion depth | T3 | T3 | T3 | T3 | T3 | T3 | T3 | T3 | T3 | T3 |
| TNM stage | IIIB | IIIB | IIIB | IIIB | IIIB | IIA | IIB | IIA | IIB | IIA |
| Number of lymph node dissection | 11 | 36 | 20 | 22 | 25 | 26 | 22 | 15 | 19 | 28 |
| Lymph node metastasis | 1 | 6 | 1 | 1 | 3 | 0 | 0 | 0 | 0 | 0 |
RNA-seq, transcriptome sequencing; ESCC, esophageal squamous cell carcinoma; TNM, tumor-node-metastasis.

Identification of GAS6-AS1 as a lymph node metastasis-related lncRNA via RNA-seq
Analysis of the intersection of the two sets of data was visualized through a Venn diagram, and several important lncRNAs were identified. In order to verify correlation between qPCR and RNA-seq data, we analyzed and compared ESCC tissues with and without lymph node metastasis. Compared to ESCC tissues without lymph node metastasis, GAS6-AS1 in ESCC tissues with lymph node metastasis was significantly increased (Figure 2A). There was no difference in the number of lymph nodes dissection by surgery between the two groups, excluding the difference in lymph node metastasis caused by the number of lymph nodes cleaned by surgery (Figure 2B). Further data analysis suggested that in the subgroup of primary tumor staging T3 group, ESCC tissue with lymph node metastasis was significantly higher than no lymph node metastasis tissue (Figure 2C). Similarly, there was no difference in the number of lymph nodes dissection by surgery between the two groups of patients (Figure 2D). Next, we used the KM survival curve to analyze OS and DFS data. We used median expression cutoff values to show that high expression levels of GAS6-AS1 were negatively correlated with DFS (Figure 2E) and OS (Figure 2F). We further analyzed whether there were differences in prognosis among patients with or without lymph node metastasis, and the results showed that the group with high expression of GAS6-AS1 and lymph node metastasis (High GAS6-AS1, LN+) had the worst prognosis. In the low expression GAS6-AS1 group, there was no difference in DFS (Figure 2G) and OS (Figure 2H) between lymph node metastasis and no metastasis groups (low GAS6-AS1, LN+vs. low GAS6-AS1, LN−). Overall, the data showed that the expression level of GAS6-AS1 is closely related to poor prognosis in these patients, especially when combined with lymph node metastasis.
GAS6-AS1 is highly expressed in ESCC issues and correlated with lymph node metastasis
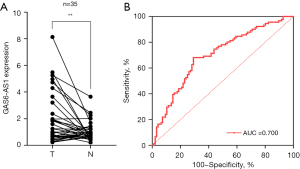
Subsequently, based on the Venn diagram intersection data, we identified several important lncRNAs, including GAS6-AS1, for which currently there is no relevant research related to ESCC. By qRT-PCR analysis of 187 ESCC tissues, we divided the GAS6-AS1 expression into a high expression group and a low expression group based on the median expression level of GAS6-AS1. We used χ2 tests to show that increased expression of GAS6-AS1 in ESCC was significantly associated with poor tumor differentiation grade (P=0.028) and was associated with tumor-node-metastasis (TNM) staging (III + IV vs. I + II; P<0.001) and primary tumor staging (T3 + T4 vs. T1 + T2; P=0.007). Importantly, increased expression of GAS6-AS1 was associated with lymph node metastasis (P<0.001). However, this is not related to the patient’s age and gender (Table 2). We further conducted univariate and multivariable logistic regression analysis to determine whether GAS6-AS1 expression is related to lymph node metastasis. The results of univariate analysis showed that there were statistical differences in primary tumor stage, tumor differentiation, and expression of GAS6-AS1. We further incorporated the above factors into multivariate analysis, and the results showed that the expression of GAS6-AS1 can serve as an independent risk factor for lymph node metastasis in ESCC [hazard ratio (HR) =4.483; 95% confidence interval (CI): 2.425–8.286; P<0.001; Table 3]. We further expanded our research data by qRT-PCR analysis of 35 paired ESCC and normal tissues, and the results showed an increase in GAS6-AS1 expression compared to paired normal esophageal epithelium (Figure 3A). Next, we analyzed whether GAS6-AS1 expression has the ability to predict lymph node metastasis. The ROC curve analysis showed that the AUC was 0.700 (P<0.001) (Figure 3B), suggesting that it is an effective indicator for predicting lymph node metastasis of ESCC, which indicated that it can be used as a potential biomarker for local regional metastasis in ESCC.
Table 2
| Clinical parameters | High expression (n=93) | Low expression (n=94) | P value |
|---|---|---|---|
| Gender | 0.068 | ||
| Male | 41 | 54 | |
| Female | 52 | 40 | |
| Age (years) | 0.407 | ||
| <60 | 19 | 24 | |
| ≥60 | 74 | 70 | |
| Differentiation grade | 0.028 | ||
| Poor | 40 | 26 | |
| Well/moderate | 53 | 68 | |
| TNM stage | <0.001 | ||
| I/II stage | 33 | 66 | |
| III/IV stage | 60 | 28 | |
| Primary tumor invasion depth | 0.007 | ||
| T1/T2 | 35 | 54 | |
| T3/T4 | 58 | 40 | |
| Lymph node metastasis | <0.001 | ||
| Negative | 31 | 65 | |
| Positive | 62 | 29 | |
ESCC, esophageal squamous cell carcinoma; TNM, tumor-node-metastasis.
Table 3
| Clinical parameters | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | ||
| Gender | 0.98 | 0.553–1.74 | 0.46 | – | – | – | |
| Age | 1.005 | 0.978–1.034 | 0.71 | – | – | – | |
| Differentiation grade | 3.884 | 2.045–7.378 | <0.001 | 3.651 | 1.826–7.302 | <0.001 | |
| Primary tumor invasion depth | 2.695 | 1.49–4.873 | 0.001 | 2.343 | 1.214–4.52 | 0.011 | |
| GAS6-AS1 expression | 4.483 | 2.425–8.286 | <0.001 | 3.723 | 1.937–7.154 | <0.001 | |
HR, hazard ratio; CI, confidence interval.
GAS6-AS1 is a crucial lncRNA for the growth and metastasis of ESCC cells
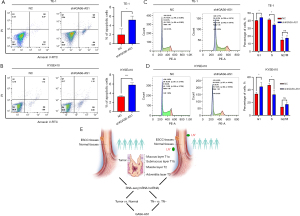
To confirm the role of GAS6-AS1 in the progression and metastasis of ESCC, we first detected the expression of GAS6-AS1 in two ESCC cell lines and one normal esophageal epithelial cell line. The results of qPCR showed that compared with the normal esophageal epithelial cell line (HET-1A), the expression of GAS6-AS1 in two ESCC cell lines was significantly increased (Figure 4A). We then used three independent shRNAs to silence GAS6-AS1 in KYSE410 and TE-1 cells (Figure 4B,4C), and the shRNA3, with better knockout effect, was selected for subsequent experiments. Related to the growth of ESCC cells, the CCK-8 assays showed that knocking down GAS6-AS1 significantly inhibited the growth of TE-1 and KYSE410 cells (Figure 4D,4E). Colony formation assays showed that knocking down the expression of GAS6-AS1 significantly reduced the colony number and size compared to the NC group (Figure 4F,4G). To confirm the role of GAS6-AS1 in the metastasis of ESCC, we used shRNA interference to knock down the expression of GAS6-AS1 in TE-1 and KYSE410, and further demonstrated through wound healing assays that GAS6-AS1 silencing inhibited cellular motility (Figure 4H,4I). Then, we established an animal xenograft model, and used the KYSE410 cell line stably silenced GAS6-AS1 or NC. In vivo experiments revealed that the tumor growth was reduced in the KYSE410-shGAS6-AS1 cell group compared with the NC group (Figure 4J), and the pictures of the isolated xenograft tumor showed that the tumors were much smaller and lighter in KYSE410-shGAS6-AS1-transplanted mice than those in the control group (Figure 4K).

The effects of GAS6-AS1 knockdown on cell promotion, apoptosis, and cell cycle
In order to further investigate the impact of GAS6-AS1 on apoptosis and cell cycle in ESCC, we used shRNA interference TE-1 and KYSE410 to knockdown the expression of GAS6-AS1, and analyzed it using flow cytometry. The results showed that knocking down GAS6-AS1 resulted in an increased apoptosis rate in KYSE410 and TE-1 cells compared to control cells. In addition, fluorescence-activated cell sorting (FACS) analysis was conducted to detect cell apoptosis (Figure 5A,5B). Cell cycle analysis showed that inhibiting the expression of GAS6-AS1 resulted in cell cycle arrest in the G1 phase and reduced the proportion of TE-1 and KYSE410 cells in the S phase (Figure 5C,5D). These results suggest that interfering with GAS6-AS1 may reduce cell proliferation by inhibiting cell cycle and promoting cell apoptosis.
Discussion
ESCC is a common tumor with a high degree of malignancy and a high risk of recurrence. Even in early ESCC, lymph node metastasis may easily occur, leading to poor prognosis for tumor patients (33). To study lymph node metastasis in ESCC, it is crucial to explore effective early predictive indicators and develop effective strategies to prevent recurrence and metastasis of ESCC. Therefore, it is essential to study the molecular mechanisms of biological processes related to lymph node metastasis in ESCC. Multiple lncRNAs have been reported to be associated with the growth and metastasis of ESCC, which might be potential biomarkers and therapeutic targets (34). For example, research has shown that lncRNA UCA1 is significantly upregulated in esophageal cancer tissues compared to adjacent normal tissues (35), and another suggested that LINC00680 promotes ESCC as a carcinogenic lncRNA and is associated with poor prognosis of ESCC (36). In addition, lncRNA ZFAS1 was identified as an oncogene upregulated in ESCC tissues, which was related to proliferation, migration, invasion, and tumor growth in vitro and induced apoptosis of ESCC cells (37).
A study has suggested that lncRNA can serve as a potential biomarker, such as lncRNA PCAT-1, may consider as a candidate prognostic biomarker for ESCC (38). However, the relationship between lymph node metastasis in ESCC and lncRNA is still unclear, and reports are scarce on whether lncRNA can predict lymph node metastasis in ESCC. Our novel findings are summarized in Figure 5E. These findings provided a significant method for screening lncRNA related to lymph node metastasis. In this study, we intersected the RNA-seq data analysis of 10 pairs of ESCC tissues with primary tumor stage T3 and their paired normal epithelium, as well as a T3N+ group and a T3N− group, to identify whether lncRNA were significantly altered with lymph node metastasis in ESCC. Similar comparative studies using microarray analysis found significant differences in the expression of 62 miRNAs in ESCC samples with lymph node metastasis compared to samples without lymph node metastasis (39). The proportion of lymph node metastasis dramatically increases after primary tumor stage T3, but there are significant differences among different patients (40). Through intersection and comparison, we may be able to identify key lncRNA that differ from lymph node metastasis in different populations. To our knowledge, this study is the first comprehensive sequencing of transcriptome in ESCC with or without lymph node metastasis at T3 stage, which is crucial for us to elucidate lymph node metastasis of ESCC at the transcriptome level. In this study, through comparison and intersection, we identified many DEGs, including some reported lncRNA, such as LINC02381, which have been confirmed to be related to the growth and apoptosis of breast cancer (24). SIM2, mRNA related to esophageal cancer and its clinical implications have been confirmed to be associated with the growth and metastasis of esophageal cancer (26). KEGG and GO analysis showed that the two DEGs were significantly enriched in cell related processes such as cell differentiation, development, death, adhesion, migration, and focal adhesion, which is closely related to the metastasis of ESCC (41). The results of qRT-PCR showed that the expression of lncRNA GAS6-AS1 is elevated in most ESCC tissues, which is consistent with the expression of most tumors, including colorectal cancer (42) and leukemia (22). Although GAS6-AS1 has been shown to have a promoting effect on tumor growth and metastasis in multiple cancer species (42), to our knowledge, there are currently no reports on the relationship between GAS6-AS1 with ESCC, especially lymph node metastasis. The relationship between lncRNA and lymph node metastasis has been reported in the literature, as the first report that lncRNA is involved in lymphatic vessel and lymph node metastasis ESCC, which may provide new insights into potential mechanisms of lymph node metastasis (16).
However, the clinical relationship between lncRNA and lymph node metastasis in ESCC is not yet clear; this is the first report that the expression level of GAS6-AS1 expression in ESCC tissue with lymph node metastasis was increased, especially in the T3 stage of the primary tumor. In addition, GAS6-AS1 is significantly elevated in ESCC tissue and paired normal esophageal epithelium, and its expression is closely related to tumor differentiation, TNM staging, T staging, and lymph node metastasis, highlighting its important role in the progression of ESCC, which is similar to clinical data of lung cancer (26). The high expression level of GAS6-AS1 in ESCC reduces the OS and DFS of patients, and especially when accompanied by lymph node metastasis, the prognosis is the worst. Next, we conducted univariate and multivariate logistic regression analyses on GAS6-AS1 expression, which is an independent risk factor for lymph node metastasis of ESCC. Next, we used ROC curve analysis to show that it is an effective indicator for predicting lymph node metastasis of ESCC. Our research shows that GAS6-AS can be used as a new potential marker for predicting lymph node metastasis of ESCC. Our study showed a significantly increased GAS6-AS1 expression in ESCC cell lines, which is consistent with the reported in lung cancer (20). In conclusion, the lncRNA GAS6-AS1 can be used as an independent risk factor for ESCC lymph node metastasis and an effective biomarker for its prediction, which may be highlighted as a promising oncogene of clinical value.
To demonstrate that GAS6-AS1 can affect the growth and metastasis of ESCC, we conducted in vivo and in vitro experiments, and the results showed that knockdown of GAS6-AS1 inhibits the growth and metastasis of ESCC in vitro, as well as the growth of tumor formation in nude mice in vivo. Subsequent studies showed that knocking down GAS6-AS1 reduced cell proliferation, and flow cytometry demonstrated that TE-1 and KYSE410 cells with shRNA had a clear cell cycle rest in the G1-S phase and the population of cells in the S phase was decreased; knocking down GAS6-AS1 also affected the apoptosis of TE-1 and KYSE410 cells. These findings confirm the functional role of GAS6-AS1 in cell growth and apoptosis, which are consistent with those of studies on other tumors (19,20).
To our knowledge, there are currently no articles reporting the mechanism of action of GAS6-AS1 in ESCC, which may be different from the mechanism of other tumors. A previous study has shown that GAS6-AS1 may affect tumor progression by regulating the expression of GAS6 (19). In addition, this type of lncRNA can serve as a sponge for miR-24-3p (26) to regulate GIMAP6. GAS6-AS1 functions as ceRNA that participates in the progression of malignant tumors, which may be the most important signaling pathway. Therefore, the interaction between GAS6-AS1 and miRNA is the most likely pathway for GAS6-AS1 to participate in tumor development. Further, the latest research shows that the targeted therapy of GAS6-AS1/YBX1/MYC axis inhibits AML cell proliferation and disease progression (22).
However, we did not study the specific signaling pathway of GAS6-AS1 affecting lymph node metastasis of ESCC in this paper. Due to the lack of sufficient evidence of specific mechanism research, we still do not know the role of GAS6-AS1, which needs to be further explored. Research of the signaling pathway will improve our understanding of its role in the progression of ESCC.
Conclusions
Our study reported the comprehensive sequencing of transcriptome in ESCC tissues with or without lymph node metastasis in T3 phase for the first time. The lncRNA GAS6-AS1 obtained from sequencing analysis can be used as an independent risk factor for ESCC lymph node metastasis and an effective biomarker for its prediction, and revealed the in vivo and in vitro effects of GAS6-AS1 on the growth and metastasis of ESCC. It has been highlighted as a promising oncogene of clinical value and more work is needed to uncover the mechanisms of GAS6-AS1.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR and ARRIVE reporting checklists. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-798/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-798/dss
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-798/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-798/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study involving human experiments was approved by the Medical Ethics Committee of the Gaozhou People’s Hospital (No. GYLLYJ-2022086). Informed consent was taken from all the patients. The animal experiments were approved by the Ethics Committee of the Gaozhou People’s Hospital (No. GYLLYJ-2022109), in compliance with national guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Global Burden of Disease Cancer Collaboration. The Global Burden of Cancer 2013. JAMA Oncol 2015;1:505-27. [Crossref] [PubMed]
- Chen W, Zheng R, Zhang S, et al. Cancer incidence and mortality in China, 2013. Cancer Lett 2017;401:63-71. [Crossref] [PubMed]
- Zeng H, Zheng R, Guo Y, et al. Cancer survival in China, 2003-2005: a population-based study. Int J Cancer 2015;136:1921-30. [Crossref] [PubMed]
- Njei B, McCarty TR, Birk JW. Trends in esophageal cancer survival in United States adults from 1973 to 2009: A SEER database analysis. J Gastroenterol Hepatol 2016;31:1141-6. [Crossref] [PubMed]
- Chau I, Norman AR, Cunningham D, et al. Multivariate prognostic factor analysis in locally advanced and metastatic esophago-gastric cancer--pooled analysis from three multicenter, randomized, controlled trials using individual patient data. J Clin Oncol 2004;22:2395-403. [Crossref] [PubMed]
- Kawahara K, Maekawa T, Okabayashi K, et al. The number of lymph node metastases influences survival in esophageal cancer. J Surg Oncol 1998;67:160-3. [Crossref] [PubMed]
- Zheng H, Tang H, Wang H, et al. Nomogram to predict lymph node metastasis in patients with early oesophageal squamous cell carcinoma. Br J Surg 2018;105:1464-70. [Crossref] [PubMed]
- Tachimori Y, Nagai Y, Kanamori N, et al. Pattern of lymph node metastases of esophageal squamous cell carcinoma based on the anatomical lymphatic drainage system. Dis Esophagus 2011;24:33-8. [Crossref] [PubMed]
- Baba H, Kanda M, Sawaki K, et al. SLC7A9 as a Potential Biomarker for Lymph Node Metastasis of Esophageal Squamous Cell Carcinoma. Ann Surg Oncol 2022;29:2699-709. [Crossref] [PubMed]
- Ghorbian S, Ardekani AM. Non-Invasive Detection of Esophageal Cancer using Genetic Changes in Circulating Cell-Free DNA. Avicenna J Med Biotechnol 2012;4:3-13. [PubMed]
- Stark R, Grzelak M, Hadfield J. RNA sequencing: the teenage years. Nat Rev Genet 2019;20:631-56. [Crossref] [PubMed]
- Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007;129:1311-23. [Crossref] [PubMed]
- Yuan K, Lan J, Xu L, et al. Long noncoding RNA TLNC1 promotes the growth and metastasis of liver cancer via inhibition of p53 signaling. Mol Cancer 2022;21:105. [Crossref] [PubMed]
- Ramli S, Sim MS, Guad RM, et al. Long Noncoding RNA UCA1 in Gastrointestinal Cancers: Molecular Regulatory Roles and Patterns, Mechanisms, and Interactions. J Oncol 2021;2021:5519720. [Crossref] [PubMed]
- Wang Y, Zhang W, Liu W, et al. Long Noncoding RNA VESTAR Regulates Lymphangiogenesis and Lymph Node Metastasis of Esophageal Squamous Cell Carcinoma by Enhancing VEGFC mRNA Stability. Cancer Res 2021;81:3187-99. [Crossref] [PubMed]
- Chen C, Zheng H, Luo Y, et al. SUMOylation promotes extracellular vesicle-mediated transmission of lncRNA ELNAT1 and lymph node metastasis in bladder cancer. J Clin Invest 2021;131:e146431. [Crossref] [PubMed]
- Sadeghpour S, Ghorbian S. Evaluation of the potential clinical prognostic value of lncRNA-BANCR gene in esophageal squamous cell carcinoma. Mol Biol Rep 2019;46:991-5. [Crossref] [PubMed]
- Zhang P, Dong Q, Zhu H, et al. Long non-coding antisense RNA GAS6-AS1 supports gastric cancer progression via increasing GAS6 expression. Gene 2019;696:1-9. [Crossref] [PubMed]
- Luo J, Wang H, Wang L, et al. lncRNA GAS6-AS1 inhibits progression and glucose metabolism reprogramming in LUAD via repressing E2F1-mediated transcription of GLUT1. Mol Ther Nucleic Acids 2021;25:11-24. [Crossref] [PubMed]
- Rankin EB, Fuh KC, Castellini L, et al. Direct regulation of GAS6/AXL signaling by HIF promotes renal metastasis through SRC and MET. Proc Natl Acad Sci U S A 2014;111:13373-8. [Crossref] [PubMed]
- Zhou H, Liu W, Zhou Y, et al. Therapeutic inhibition of GAS6-AS1/YBX1/MYC axis suppresses cell propagation and disease progression of acute myeloid leukemia. J Exp Clin Cancer Res 2021;40:353. [Crossref] [PubMed]
- Ghafouri-Fard S, Khoshbakht T, Taheri M, et al. A review on the role of GAS6 and GAS6-AS1 in the carcinogenesis. Pathol Res Pract 2021;226:153596. [Crossref] [PubMed]
- Wu G, Ma Z, Cheng Y, et al. Targeting Gas6/TAM in cancer cells and tumor microenvironment. Mol Cancer 2018;17:20. [Crossref] [PubMed]
- Guo X, Li H, Zhang M, et al. LncRNA GAS6 antisense RNA 1 facilitates the tumorigenesis of clear cell renal cell carcinoma by regulating the AMP-activated protein kinase/mTOR signaling pathway. Oncol Lett 2021;22:727. [Crossref] [PubMed]
- Wang Y, Ma M, Li C, et al. GAS6-AS1 Overexpression Increases GIMAP6 Expression and Inhibits Lung Adenocarcinoma Progression by Sponging miR-24-3p. Front Oncol 2021;11:645771. [Crossref] [PubMed]
- Tamaoki M, Komatsuzaki R, Komatsu M, et al. Multiple roles of single-minded 2 in esophageal squamous cell carcinoma and its clinical implications. Cancer Sci 2018;109:1121-34. [Crossref] [PubMed]
- Gene Ontology Consortium. going forward. Nucleic Acids Res 2015;43:D1049-56. [Crossref] [PubMed]
- Bolger GB. The cAMP-signaling cancers: Clinically-divergent disorders with a common central pathway. Front Endocrinol (Lausanne) 2022;13:1024423. [Crossref] [PubMed]
- Piazza GA, Ward A, Chen X, et al. PDE5 and PDE10 inhibition activates cGMP/PKG signaling to block Wnt/β-catenin transcription, cancer cell growth, and tumor immunity. Drug Discov Today 2020;25:1521-7. [Crossref] [PubMed]
- Sharma U, Kaur Rana M, Singh K, et al. LINC00324 promotes cell proliferation and metastasis of esophageal squamous cell carcinoma through sponging miR-493-5p via MAPK signaling pathway. Biochem Pharmacol 2023;207:115372. [Crossref] [PubMed]
- Taniguchi N, Kizuka Y. Glycans and cancer: role of N-glycans in cancer biomarker, progression and metastasis, and therapeutics. Adv Cancer Res 2015;126:11-51. [Crossref] [PubMed]
- Lagergren J, Smyth E, Cunningham D, et al. Oesophageal cancer. Lancet 2017;390:2383-96. [Crossref] [PubMed]
- Luo XJ, He MM, Liu J, et al. LncRNA TMPO-AS1 promotes esophageal squamous cell carcinoma progression by forming biomolecular condensates with FUS and p300 to regulate TMPO transcription. Exp Mol Med 2022;54:834-47. [Crossref] [PubMed]
- Aalijahan H, Ghorbian S. Clinical Application of Long Non-Coding RNA-UCA1 as a Candidate Gene in Progression of Esophageal Cancer. Pathol Oncol Res 2020;26:1441-6. [Crossref] [PubMed]
- Xue ST, Zheng B, Cao SQ, et al. Long non-coding RNA LINC00680 functions as a ceRNA to promote esophageal squamous cell carcinoma progression through the miR-423-5p/PAK6 axis. Mol Cancer 2022;21:69. [Crossref] [PubMed]
- Li Z, Qin X, Bian W, et al. Exosomal lncRNA ZFAS1 regulates esophageal squamous cell carcinoma cell proliferation, invasion, migration and apoptosis via microRNA-124/STAT3 axis. J Exp Clin Cancer Res 2019;38:477. [Crossref] [PubMed]
- Razavi M, Ghorbian S. Up-regulation of long non-coding RNA-PCAT-1 promotes invasion and metastasis in esophageal squamous cell carcinoma. EXCLI J 2019;18:422-8. [PubMed]
- Xu Y, Sun D, Zhang X, et al. Microarray analysis of miRNA based on the regional lymph node metastasis status of esophageal squamous cell carcinoma. Transl Cancer Res 2021;10:273-87. [Crossref] [PubMed]
- Deng HY, Wang ZQ, Wang YC, et al. Oesophageal adenocarcinoma has a higher risk of lymph node metastasis than squamous cell carcinoma: a propensity score-matched study. Eur J Cardiothorac Surg 2017;52:958-62. [Crossref] [PubMed]
- Guo W, Tan F, Huai Q, et al. Comprehensive Analysis of PD-L1 Expression, Immune Infiltrates, and m6A RNA Methylation Regulators in Esophageal Squamous Cell Carcinoma. Front Immunol 2021;12:669750. [Crossref] [PubMed]
- Chen Q, Zhou L, Ma D, et al. LncRNA GAS6-AS1 facilitates tumorigenesis and metastasis of colorectal cancer by regulating TRIM14 through miR-370-3p/miR-1296-5p and FUS. J Transl Med 2022;20:356. [Crossref] [PubMed]


