
LncRNA PCAT6 is a predictor of poor prognosis of colorectal cancer
Highlight box
Key findings
• Prostate cancer-associated transcript 6 (PCAT6) is a possible prognostic marker of low survival rate for patients with colorectal cancer (CRC).
What is known and what is new?
• CRC, as a common malignant tumor in the gastrointestinal tract/intestine or large intestine, is a major threat to physical health.
• In this study, CRC-related RNA sequencing (RNA-Seq) data is acquired from The Cancer Genome Atlas (TCGA) for further accessing the prognostic value of the long non-coding RNA (lncRNA) PCAT6 expression. It is found that CRC patients with PCAT6 have significantly higher expression.
What is the implication, and what should change now?
• This study provides promising insights to exhume the clinical pathological significance and molecular etiology of CRC.
Introduction
Colorectal cancer (CRC) is a common form of gastrointestinal malignancy that accounts for roughly 9,000,000 deaths throughout the world each year, making it the fourth leading cause of cancer-associated death. Rates of CRC are rising both due to the general aging of the global population and to a number of modifiable risk factors such as poor diet, obesity, smoking, and a lack of exercise (1). CRC tumors arise when healthy cells of the colonic epithelium mutate and form benign adenomas that in turn progress to a malignant state (2). Quantitative analyses suggest that early CRC-predisposing mutations occur in stem cells, while the acquisition of malignant and metastatic phenotypes can be a prolonged process that can take upwards of 10 years to complete (3). As such, diagnosing CRC in its early stages is essential yet clinically challenging. Roughly 43% and 25% of CRC patients exhibit liver and liver/lung metastases, respectively, and for individuals with stage IV disease, the 5-year overall survival (OS) rate is under 10% (4,5). The primary treatment for CRC patients is surgical tumor resection supplemented by chemotherapeutic and immunotherapeutic treatment. However, these treatments are of limited value for individuals with advanced disease. Several promising biomarkers have been tested as potential predictors of CRC onset or progression in clinical contexts (6), including carcinoembryonic antigen (CEA) and carbohydrate antigen 199 (CA199) (7), yet their utility remains a matter of some controversy. There is thus an urgent need to develop novel diagnostic and therapeutic biomarkers for this cancer type. Research has shown that in the advanced disease setting, data highlight the potential of circulating tumor DNA (ctDNA) levels as a prognostic marker and as an early indicator of treatment response. ctDNA assessment can complement standard tissue-based testing for molecular characterisation, with the added ability to monitor emerging mutations under the selective pressure of targeted therapy, they provide an overview of the evidence supporting the use of ctDNA in CRC (8,9).
prostate cancer-associated transcript 6 (PCAT6; PCAN-R1, ncRNA-a2, KDM5B-AS1) is a recently described long non-coding RNA (lncRNA) associated with oncogenic processes (10). PCAT6 is a 764 bp lncRNA encoded in an intergenic region on chromosome 1q32.1, where it flanks the histone demethylase JARID1B/KDAM 5B gene. In prostate tumor cells, this lncRNA was shown to induce colony formation and keratinocyte proliferation (11). Its upregulation has also been reported in a variety of tumors including bladder, ovarian, lung, and gastric cancers, as well as in osteosarcoma, hepatocellular carcinoma (HCC), and glioblastoma (12). PCAT6 can increase the proliferative, migratory, and angiogenic activity of triple-negative breast cancer (TNBC) cells in vitro and in vivo, and vascular endothelial growth factor (VEGF) derived from M2 macrophages can stimulate PCAT6 upregulation to promote angiogenic activity in the context of TNBC (13). PCAT6 upregulation in HCC has been shown to be associated with worse overall survival (OS) and disease-free survival, in addition to enhancing the proliferative activity and survival of HCC cells, whereas the knockdown of this lncRNA induced cell cycle arrest and apoptotic death in these same tumor cells (14). In bladder cancer tissues, PCAT6 levels have been reported to be higher than in paracancerous healthy tissues, with bladder cancer patients exhibiting increased serum PCAT6 levels as compared to healthy controls suggesting that it may represent a valuable diagnostic biomarker for this cancer type (15). However, there have been few studies to date assessing the relationship between PCAT6 and CRC.
As such, we sought to establish the relationship between PCAT6 and CRC, and to assess the prognostic relevance of this lncRNA based on an analysis of CRC patient data compiled within The Cancer Genome Atlas (TCGA). To that end, we compared PCAT6 expression in CRC tumors and normal tissue samples in the TCGA database, and examined correlations between PCAT6 levels and patient prognosis. Moreover, a gene set enrichment analysis (GSEA) technique was used to identify PCAT6-related biological processes with potential relevance to the progression of CRC.
In this study, we found PCAT6 upregulation to be evident in CRC patients, associated with worse CRC patient prognostic outcomes, and related to mechanistic functions including base excision repair, the G2/M DNA damage checkpoint, cellular senescence, chromatin-modifying enzymes, DNA methylation, and RNA-mediated gene silencing via a GSEA approach. As such, PCAT6 may represent a valuable biomarker for the diagnosis and prognostic assessment of individuals with CRC. We present this article in accordance with the REMARK reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-910/rc).
Methods
TCGA data collection
Level 3 RNA sequencing (RNA-Seq) data in the HTSeq-FPKM format with corresponding clinical details were downloaded from the TCGA (https://portal.gdc.cancer.gov/) CRC colon adenocarcinoma (COAD) and rectal adenocarcinoma (READ) projects, with any samples for which clinical information was missing being discarded, yielding 644 samples for subsequent analysis (Table 1). The detailed clinicopathologic information such as patient age, pathologic stage, neoplasm type (READ vs. COAD), height, weight, gender, race, history of colon polyps, colon polyps present, and lymphatic invasion status were assessed, as were CEA levels, residual tumor status, and tumor-node-metastasis (TNM) staging. All data were obtained from a publicly accessible database, and no direct human or animal research was conducted by the authors of this study. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Table 1
| Clinical characteristic | Values (n=644) |
|---|---|
| T stage, n (%) | |
| T1 | 20 (3.1) |
| T2 | 111 (17.3) |
| T3 | 436 (68.0) |
| T4 | 74 (11.5) |
| N stage, n (%) | |
| N0 | 368 (57.5) |
| N1 | 153 (23.9) |
| N2 | 119 (18.6) |
| M stage, n (%) | |
| M0 | 475 (84.2) |
| M1 | 89 (15.8) |
| Pathologic stage, n (%) | |
| Stage I | 111 (17.8) |
| Stage II | 238 (38.2) |
| Stage III | 184 (29.5) |
| Stage IV | 90 (14.4) |
| CEA level, n (%) | |
| ≤5 | 261 (62.9) |
| >5 | 154 (37.1) |
| Lymphatic invasion, n (%) | |
| No | 350 (60.1) |
| Yes | 232 (39.9) |
| History of colon polyps, n (%) | |
| No | 377 (67.9) |
| Yes | 178 (32.1) |
| Colon polyps present, n (%) | |
| No | 224 (69.3) |
| Yes | 99 (30.7) |
| Neoplasm type, n (%) | |
| Colon adenocarcinoma | 478 (74.2) |
| Rectum adenocarcinoma | 166 (25.8) |
| Gender, n (%) | |
| Female | 301 (46.7) |
| Male | 343 (53.3) |
| Race, n (%) | |
| Asian | 12 (3) |
| Black or African American | 69 (17.5) |
| White | 313 (79.4) |
| Age (years), median [IQR] | 68 [58–76] |
TCGA, The Cancer Genome Atlas; T, primary tumor; N, lymph node metastasis; M, distant metastasis; CEA, carcinoembryonic antigen; IQR, interquartile range.
GSEA
GSEA approaches represent a computational means of establishing whether a given set of genes differ significantly between a given set of biological states (16). GSEA analyses were performed based on patients in the TCGA COAD and READ datasets with low and high PCAT6 expression levels using the clusterPorfiler package (17). Ordered gene lists were generated based on the strength of individual correlations with PCAT6 in this analysis, with GSEA being conducted to clarify differences in survival outcomes between patients with low and high PCAT6 expression levels. A preliminary GSEA version was utilized for data analysis (18). Analyses were conducted with 10,000 genome permutations, with PCAT6 expression levels as a phenotypic label. Pathway enrichment was assessed according to nominal P values and normalized enrichment score (NES) value.
Single-sample GSEA (ssGSEA)-based immune cell analysis
A ssGSEA analysis was used to evaluate immune cell infiltration into CRC tumors with the R gene set variation analysis (GSVA) package (19). This approach allowed for GSEA-based analyses of 24 different immune cell populations including natural killer (NK) cells, eosinophils, CD8+ T cells, B cells, Th17 cells, T cells, gamma delta T (Tgd) cells, immature dendritic cells (iDCs), mast cells, eosinophils, Th1 cells, plasmacytoid DCs (pDCs), neutrophils, activated DCs (aDCs), DCs, T helper cells, NK CD56dim cells, T follicular helper (Tfh) cells, T effector memory (Tem) cells, macrophages, Th2 cells, central memory T (Tcm) cells, and cytotoxic cells. Based upon characteristic gene expression patterns for these cell types (20), their relative enrichment in each tumor sample was established. Spearman correlation analyses and Wilcoxon rank-sum tests were then used to assess correlations between PCAT6 expression and such immune cell infiltration in patients with different levels of PCAT6 expression.
RNA extraction and quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total cellular RNA was first extracted with Trizol Reagent (Magen Biotech Co., Ltd., Guangzhou, China); then reverse transcribed into complementary DNA (cDNA) with HiScript ‖Q RT SuperMix (Vazyme Biotech Co., Ltd., Nanjing, China); then RT-PCR kit (AceQ qPCR SYBR Green Master Mix, Q111-02) and LightCycler 480 PCR system (Roche) instrument, GAPDH was used as an internal reference; finally, the 2−ΔΔCt method was used to calculate the relative expression of lncRNA PCAT6. The primer base sequences (forward/reverse) used were as follows: PCAT6: TGCTTCTACCACCACCCTTC/TTCACAGGGGACATCTGACA; GAPDH: CAGGAGGCATTGCTGATGAT/GAAGGCTGGGGCTCATTT. Data are presented as three independent experiments.
Statistical analysis
R (3.6.3) was used to analyze all data in the present study (https://www.xiantao.love/). Associations between PCAT6 expression levels and clinicopathological variables were assessed via Wilcoxon rank-sum tests and logistic regression analyses, while the associations between patient OS and specific clinicopathological variables were assessed through Kaplan-Meier and Cox regression analyses. The relationship between PCAT6 expression levels, other variables of interest, and patient survival was assessed via a multivariate Cox analysis. Median values were used to stratify patients into subgroups with low and high levels of PCAT6 expression. P<0.05 was the threshold of significance.
Results
Patients and samples
In total, 644 patients were identified for inclusion in this study (343 male, 301 female, Table 1). Of these patients, 232 (39.9%) exhibited lymphatic invasion, 178 (32.1%) had a pre-treatment history of colon polyps, and 99 (30.7%) exhibited colonic polyps. Patients with stage I, II, III, and IV disease accounted for 17.8% (n=111), 38.2% (n=238), 29.5% (n=184), and 14.4% (n=90) of the overall cohort, respectively, with 478 (74.2%) and 166 (25.8%) respective cases of COAD and READ. With respect to staging, 3.1% (n=20), 17.3% (n=111), 68% (n=436), and 11.5% (n=74) of patients had T1 (primary tumor), T2, T3, and T4 disease; 57.5% (n=368), 23.9% (n=153), and 18.6% (n=119) had N0, N1, and N2 disease; and 84.2% (n=475) and 15.8% (n=89) had M0 and M1 disease, respectively. In addition, 154 patients (37.1%) had pre-treatment CEA levels greater than five.
Assessment of PCAT6 expression and diagnostic utility in CRC
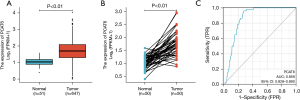
Using a Wilcoxon rank-sum test, PCAT6 expression was next compared between 647 CRC tumor samples and 51 normal tissue samples, revealing this lncRNA to be significantly upregulated in tumor tissue samples relative to healthy control tissues (P<0.01) (Figure 1A). When PCAT6 levels were compared between CRC patient tumors and matched paracancerous tissues, this lncRNA was similarly found to be upregulated in tumor tissues (P<0.01) (Figure 1B), suggesting that it may be linked to CRC development and/or progression. Receiver operating characteristic (ROC) curves were then generated to assess the diagnostic efficacy of PCAT6 based upon TCGA data, yielding an area under the ROC curve (AUC) value of 0.859 (Figure 1C), consistent with the ability of this lncRNA to reliably discriminate between tumors and healthy tissues.
The association between PCAT6 expression and CRC patient clinicopathological characteristics
Next, the relationship between PCAT6 expression levels and clinical characteristics for 647 CRC patients in the TCGA database was assessed. As shown in Figure 2, high levels of PCAT6 expression were significantly correlated with tumor N stage (N0 vs. N1 vs. N2, P<0.01), M stage (M1 vs. M0, P<0.01), pathological stage (stage I vs. stage II vs. stage III vs. stage IV, P<0.05), lymphatic invasion (yes vs. no, P<0.001), tumor type (READ vs. COAD, P<0.001), and CEA level (≤5 vs. >5, P<0.01). In univariate analyses, PCAT6 expression was associated with poor clinicopathological characteristics when using a median expression cutoff threshold of 2.5 (Table 2). Higher PCAT6 expression levels in CRC were associated with N stage [odds ratio (OR) =1.81 for N1 & N2 vs. N0, P<0.001], M stage (OR =2.07 for M1 vs. M0, P=0.002), CEA level (OR =1.88 for >5 vs. ≤5, P=0.002), lymphatic invasion (OR =1.95 for yes vs. no, P<0.001), pathological stage (OR =1.94 for stage III/IV vs. stage I/II, P<0.001), and tumor type (OR =2.14 for READ vs. COAD, P<0.001). These findings suggested that CRC patients exhibiting increased PCAT6 expression were more likely to harbor lymph node metastases and more advanced disease.
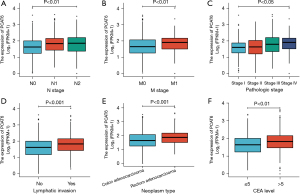
Table 2
| Characteristics | Total | Odds ratio in PCAT6 expression (95% CI) | P value |
|---|---|---|---|
| T stage (T3 & T4 vs. T1 & T2) | 641 | 1.449 (0.985–2.140) | 0.061 |
| N stage (N1 & N2 vs. N0) | 640 | 1.809 (1.319–2.488) | <0.001 |
| M stage (M1 vs. M0) | 564 | 2.065 (1.300–3.330) | 0.002 |
| CEA level (>5 vs. ≤5) | 415 | 1.884 (1.260–2.831) | 0.002 |
| Perineural invasion (yes vs. no) | 235 | 1.909 (1.057–3.499) | 0.034 |
| History of colon polyps (yes vs. no) | 555 | 0.721 (0.503–1.031) | 0.074 |
| Colon polyps present (yes vs. no) | 323 | 0.674 (0.417–1.084) | 0.105 |
| Lymphatic invasion (yes vs. no) | 582 | 1.947 (1.392–2.733) | <0.001 |
| Pathologic stage (stage III & IV vs. stage I & II) | 623 | 1.942 (1.410–2.681) | <0.001 |
| Neoplasm type (rectum adenocarcinoma vs. colon adenocarcinoma) | 644 | 2.143 (1.493–3.097) | <0.001 |
lncRNA, long non-coding RNA; PCAT6, prostate cancer-associated transcript 6; T, primary tumor, N, lymph node metastasis; M, distant metastasis, CEA, carcinoembryonic antigen.
Examination of the relationship between PCAT6 and patient survival
Kaplan-Meier survival analyses revealed CRC patients with high PCAT6 levels exhibited a worse prognosis than that of patients expressing lower levels of this lncRNA (P=0.017) (Figure 3A), with similar results being obtained when assessing the progression-free interval (P=0.001) (Figure 3B). In univariate analyses, high PCAT6 expression was associated with poorer OS [hazard ratio (HR): 1.540; 95% confidence interval (CI): 1.079–2.199; P=0.017], and other variables associated with lower survival rates included age, CEA levels, lymphatic invasion, pathological stage, and TNM stage. In multivariate analyses, an independent association between PCAT6 expression and OS was detected (HR =6.892; 95% CI: 1.713–27.727, P=0.007), with the same being true for age and M stage (Table 3).
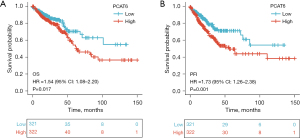
Table 3
| Characteristics | Total | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | |||
| T stage (T1 & T2 vs. T3 & T4) | 640 | 2.468 (1.327–4.589) | 0.004 | 0.258 (0.044–1.517) | 0.134 | |
| N stage (N0 vs. N1 & N2) | 639 | 2.627 (1.831–3.769) | <0.001 | 4.661 (0.804–27.013) | 0.086 | |
| M stage (M0 vs. M1) | 563 | 3.989 (2.684–5.929) | <0.001 | 5.691 (1.220–26.546) | 0.027 | |
| CEA level (≤5 vs. >5) | 414 | 2.620 (1.611–4.261) | <0.001 | 1.178 (0.296–4.687) | 0.816 | |
| Perineural invasion (no vs. yes) | 235 | 1.692 (0.907–3.156) | 0.099 | |||
| Lymphatic invasion (no vs. yes) | 581 | 2.144 (1.476–3.114) | <0.001 | 0.958 (0.295–3.106) | 0.942 | |
| Pathologic stage (stage I & II vs. stage III & IV) | 622 | 2.988 (2.042–4.372) | <0.001 | 2.085 (0.505–8.617) | 0.310 | |
| Colon polyps present (no vs. yes) | 323 | 1.250 (0.743–2.103) | 0.401 | |||
| History of colon polyps (no vs. yes) | 554 | 0.789 (0.496–1.257) | 0.319 | |||
| Race (Asian vs. Black or African American & White) | 394 | 0.840 (0.205–3.438) | 0.809 | |||
| Gender (female vs. male) | 643 | 1.054 (0.744–1.491) | 0.769 | |||
| Weight (≤90 vs. >90 kg) | 348 | 0.742 (0.412–1.339) | 0.322 | |||
| Height (<170 vs. ≥170 cm) | 329 | 0.779 (0.473–1.281) | 0.324 | |||
| Age (≤65 vs. >65 years) | 643 | 1.939 (1.320–2.849) | <0.001 | 9.813 (2.525–38.135) | <0.001 | |
| PCAT6 (low vs. high) | 643 | 1.540 (1.079–2.199) | 0.017 | 6.892 (1.713–27.727) | 0.007 | |
PCAT6, prostate cancer-associated transcript 6; CI, confidence interval; T, primary tumor; N, lymph node metastasis; M, distant metastasis; CEA, carcinoembryonic antigen.
GSEA-based identification of signaling pathways associated with PCAT6
Next, a GSEA approach was used to compare patterns of gene expression between CRC tumors with low and high levels of PCAT6 expression, revealing significant differences in enrichment levels for MSigDB collections (c2.cp.v7.0.symbols.gmt) [false discovery rate (FDR) <0.05, normalized P<0.05]. NES values were then used to select the most enriched signaling pathways (Figure 4 and Table 4), revealing high levels of PCAT6 expression to be associated with the enrichment of DNA methylation, RNA-mediated gene silencing, cellular senescence, base excision repair, chromatin-modifying enzyme, and G2/M DNA damage checkpoint pathways.
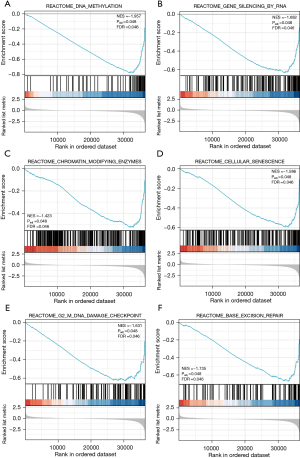
Table 4
| MSigDB collection | Gene set name | NES | Padj | FDR |
|---|---|---|---|---|
| c2.cp.v7.0.symbols.gmt | REACTOME_DNA_METHYLATION | −1.957 | 0.048 | 0.046 |
| REACTOME_GENE_SILENCING_BY_RNA | −1.682 | 0.048 | 0.046 | |
| REACTOME_CHROMATIN_MODIFYING_ENZYMES | −1.423 | 0.048 | 0.046 | |
| REACTOME_CELLULAR_SENESCENCE | −1.586 | 0.048 | 0.046 | |
| REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT | −1.631 | 0.048 | 0.046 | |
| REACTOME_BASE_EXCISION_REPAIR | −1.735 | 0.048 | 0.046 |
NES, normalized enrichment score; Padj, adjusted P value; FDR, false discovery rate.
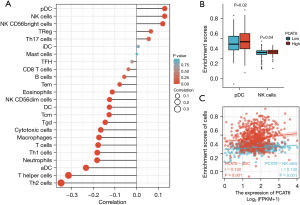
Immune cell infiltration analysis of PCAT6 in the CRC
Finally, we examined the relationship between PCAT6 expression and immune cell infiltration as quantified via an ssGSEA analysis in CRC samples through a Spearman correlation approach. This strategy revealed higher levels of PCAT6 expression to be positively correlated with pDC and NK cell infiltration (P=0.001, Figure 5).
Expression of lncRNA PCAT6 in CRC cell lines
The expression level of PCAT6 in CRC cell lines was detected by qRT-PCR. As shown in Figure 6, the expression of PCAT6 in the two CRC cell lines was higher than that in normal colon epithelial cells, and the difference was statistically significant (P<0.01). Among them, PCAT6 had the highest expression in Human Colorectal Adenocarcinoma Cells (RKO) cells, so it can be selected as a cell model for follow-up experiments to examine the regulatory role of PCAT6 in the progression of RKO cells (Figure 6).

Discussion
The lncRNA colorectal neoplasia differentially expressed (CRNDE) is considered a carcinogenic promoter in various human malignancies. LncRNAs are associated with tumorigenesis of liver cancer. LncRNA CRNDE was identified as an oncogenic lncRNA and involved in tumor growth and metastasis (21-23). A growing body of research has shown PCAT6 playing key roles in many different cancers, with the upregulation of this lncRNA being associated with worse prognostic outcomes and clinical features, underscoring its promising utility as a prognostic biomarker (24). For example, one study of osteosarcoma samples revealed higher PCAT6 expression in these tumors relative to paracancerous normal bone tissue, with such upregulation being linked to worse patient survival and a more malignant phenotype (25). Similarly, PCAT6 overexpression in cholangiocarcinoma (CCA) has been shown to be linked to the modulation of macrophage function in affected patients, suggesting that it may be a viable immunotherapeutic target in this oncogenic setting (26). However, few studies have examined the relevance of PCAT6 in CRC. This study was thus conducted to compare PCAT6 expression levels in CRC tissues and to explore its associated prognostic and therapeutic utility. Overall, we found that PCAT6 is upregulated in CRC tumors relative to healthy normal tissue levels, suggesting that it may play a tumorigenic role in affected patients.
Herein, we analyzed RNA-seq data pertaining to CRC patients derived from the TCGA database, revealing significant PCAT6 upregulation in CRC tumors relative to normal tissue samples. Such upregulation was tied to certain characteristics of advanced disease (lymphatic invasion, pathological stage, TNM stage, tumor type) and to a poorer overall patient prognosis. GSEA results revealed PCAT6 upregulation to be linked to biological activities including base excision repair, G2/M DNA damage checkpoint, cellular senescence, DNA methylation, and RNA-mediated gene silencing, all of which are linked to tumor cell invasion, proliferation, and metastasis (27-32). PCAT6 may thus represent a promising new therapeutic and prognostic target in CRC patients.
In ovarian cancer cells, PCAT6 has been found to be upregulated and to enhance migratory, invasive, and proliferative activity levels by inhibiting the tumor suppressor gene PTEN in a manner closely tied to distant metastasis or lymph node metastasis (33). Similar findings have also been reported in intrahepatic CCA, with PCAT6 upregulation being linked to more advanced disease (34). Higher PCAT6 levels have also been reported in glioblastoma cells and tissues, functioning through a positive feedback mechanism to regulate the progression of this cancer type (35). In line with these prior reports, we herein found PCAT6 to be upregulated in CRC and to be correlated with patient TNM stage, lymph node metastasis, and clinical stage, suggesting that it may be a clinically relevant indicator of CRC development and progression.
DNA methylation is a common epigenetic modification that can control associated transcriptional activity. By regulating DNA methyltransferase activity and associated enzymes or by modulating substrate availability, dietary compounds can impact DNA methylation. These epigenetic modifications are thought to be directly relevant to carcinogenic processes, and offer substantial therapeutic potential (36). Patterns of DNA methylation in thyroid cancer have been previously explored as promising prognostic and therapeutic targets with the potential to benefit patients with thyroid cancer (37). Increased signals associated with cancer gene-silencing activities for certain miRNAs are increased, enhancing cancer cell migration and proliferation (38). This suggests that the DNA methylation can influence CRC cells through pathways related to RNA-mediated gene silencing.
The present analyses further revealed close correlations between PCAT6 expression levels and immune cell infiltration in CRC tumors. Specifically, PCAT6 expression levels were moderately to strongly positively correlated with NK and pDC cell infiltration, in addition to being significantly correlated with Treg, Th17, and NK CD56bright cell infiltration. This suggests that PCAT6 may shape intratumoral immune responses in CRC. In contrast to the robust relationship observed for pDCs, aDCs were only weakly correlated with PCAT6 expression, suggesting a link between this lncRNA and tumor-associated DC polarization. Moreover, PCAT6 levels were closely correlated with expression levels of many NK cell-related markers, suggesting a link between this lncRNA and these important cytolytic cells. These correlative relationships suggest that PCAT6 can affect immune cell recruitment and infiltration in CRC tumors.
The results of this study offer new insights into the functional importance of PCAT6 in CRC. However, further work will be needed to validate and expand upon these results. For example, additional analyses of clinical factors associated with changes in PCAT6 expression levels warrant further in-depth study. In addition, the data analyzed herein were derived from multiple laboratories, potentially resulting in inconsistent sample and data processing. Moreover, the number of healthy controls in this study differed significantly from the number of CRC samples, necessitating follow-up studies in which sample sizes are more appropriately balanced. Even so, as a multi-center study, this analysis may overcome some of the limitations of single-center studies. As this was a retrospective study, data pertaining to certain interventions were missing and certain information was insufficiently specific. Further prospective work will overcome these potential analytical biases. Additionally, this was an RNA-seq based analysis, and correlations between PCAT6 expression and proteomic changes thus warrant further study to clarify the direct mechanisms whereby this lncRNA influences CRC onset and progression. Future cell line- and animal model-based studies may offer greater insight into how PCAT6 shapes to oncogenic processes highlighted in this report.
Conclusions
In summary, the results of this study suggest that the upregulation of PCAT6 is closely tied to the progression of CRC, immune cell infiltration, and worse patient survival. The high expression of lncRNA PCAT6 in CRC cell lines was demonstrated by PCR experiments. This lncRNA may thus promote dysregulated immune and inflammatory responses, driving oncogenic progression. While these results offer new insight into the pathological basis of CRC, additional prospective analyses and randomized clinical trials will be essential to further clarify the underlying molecular mechanisms and clinical relevance in patients with this deadly cancer type.
Acknowledgments
Funding: The authors would like to acknowledge the financial supports from
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-910/rc
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-910/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-910/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Dekker E, Tanis PJ, Vleugels JLA, et al. Colorectal cancer. Lancet 2019;394:1467-80. [Crossref] [PubMed]
- Binefa G, Rodríguez-Moranta F, Teule A, et al. Colorectal cancer: from prevention to personalized medicine. World J Gastroenterol 2014;20:6786-808. [Crossref] [PubMed]
- Han L, Sun Y, Lu C, et al. MiR-3614-5p Is a Potential Novel Biomarker for Colorectal Cancer. Front Genet 2021;12:666833. [Crossref] [PubMed]
- Chuang HY, Jiang JK, Yang MH, et al. Aminopeptidase A initiates tumorigenesis and enhances tumor cell stemness via TWIST1 upregulation in colorectal cancer. Oncotarget 2017;8:21266-80. [Crossref] [PubMed]
- Bora K, Bhuyan MK, Kasugai K, et al. Computational learning of features for automated colonic polyp classification. Sci Rep 2021;11:4347. [Crossref] [PubMed]
- Gires O. Lessons from common markers of tumor-initiating cells in solid cancers. Cell Mol Life Sci 2011;68:4009-22. [Crossref] [PubMed]
- Chen W, Liu Q, Tan SY, et al. Association between carcinoembryonic antigen, carbohydrate antigen 19-9 and body mass index in colorectal cancer patients. Mol Clin Oncol 2013;1:879-86. [Crossref] [PubMed]
- Loft M, To YH, Gibbs P, et al. Clinical application of circulating tumour DNA in colorectal cancer. Lancet Gastroenterol Hepatol 2023;8:837-52. [Crossref] [PubMed]
- Xue J, Prabhakaran S, Prabhakaran S, et al. The utility of ctDNA in colorectal cancer with peritoneal metastases. ANZ J Surg 2023;93:506-9. [Crossref] [PubMed]
- Prensner JR, Iyer MK, Balbin OA, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol 2011;29:742-9. [Crossref] [PubMed]
- Du Z, Fei T, Verhaak RG, et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol 2013;20:908-13. [Crossref] [PubMed]
- Wang S, Chen Z, Gu J, et al. The Role of lncRNA PCAT6 in Cancers. Front Oncol 2021;11:701495. [Crossref] [PubMed]
- Dong F, Ruan S, Wang J, et al. M2 macrophage-induced lncRNA PCAT6 facilitates tumorigenesis and angiogenesis of triple-negative breast cancer through modulation of VEGFR2. Cell Death Dis 2020;11:728. Erratum in: Cell Death Dis 2022;13:752. [Crossref] [PubMed]
- Luo Y, Lin J, Zhang Y, et al. LncRNA PCAT6 predicts poor prognosis in hepatocellular carcinoma and promotes proliferation through the regulation of cell cycle arrest and apoptosis. Cell Biochem Funct 2020;38:895-904. [Crossref] [PubMed]
- Zhang D, Du D, Yi S, et al. LncRNA PCAT6: A potential biomarker for diagnosis and prognosis of bladder cancer. Ann Diagn Pathol 2020;49:151642. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003;34:267-73. [Crossref] [PubMed]
- Barbie DA, Tamayo P, Boehm JS, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009;462:108-12. [Crossref] [PubMed]
- Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013;39:782-95. [Crossref] [PubMed]
- Zhang T, Fu MC, Tan WQ, et al. Long Non-Coding RNA CRNDE Functions as a Tumor Promoter by Modulating the ERK/MAPK Signaling Pathway in Neuroblastoma Cells. Ann Clin Lab Sci 2022;52:956-66.
- Gao P, Sun D, Guo H, et al. LncRNA CCAT2 promotes proliferation and suppresses apoptosis of colorectal cancer cells. J BUON 2020;25:1840-6.
- Tang D, Zhao L, Mu R, et al. LncRNA colorectal neoplasia differentially expressed promotes glycolysis of liver cancer cells by regulating hypoxia-inducible factor 1α. Chin J Physiol 2022;65:311-8. [Crossref] [PubMed]
- Shi SB, Cheng QH, Gong SY, et al. PCAT6 may be a new prognostic biomarker in various cancers: a meta-analysis and bioinformatics analysis. Cancer Cell Int 2021;21:370. [Crossref] [PubMed]
- Wu K, Feng Q, Li L, et al. Long-Noncoding RNA PCAT6 Aggravates Osteosarcoma Tumourigenesis via the MiR-143-3p/ZEB1 Axis. Onco Targets Ther 2020;13:8705-14. [Crossref] [PubMed]
- Tu J, Wu F, Chen L, et al. Long Non-Coding RNA PCAT6 Induces M2 Polarization of Macrophages in Cholangiocarcinoma via Modulating miR-326 and RhoA-ROCK Signaling Pathway. Front Oncol 2021;10:605877. Erratum in: Front Oncol 2021;11:680500. [Crossref] [PubMed]
- Yu J, Hua R, Zhang Y, et al. DNA hypomethylation promotes invasion and metastasis of gastric cancer cells by regulating the binding of SP1 to the CDCA3 promoter. J Cell Biochem 2020;121:142-51. [Crossref] [PubMed]
- Xu Y, Qin L, Sun T, et al. Twist1 promotes breast cancer invasion and metastasis by silencing Foxa1 expression. Oncogene 2017;36:1157-66. [Crossref] [PubMed]
- Ma C, Wang F, Han B, et al. SALL1 functions as a tumor suppressor in breast cancer by regulating cancer cell senescence and metastasis through the NuRD complex. Mol Cancer 2018;17:78. Erratum in: Mol Cancer 2022;21:90. [Crossref] [PubMed]
- Lee S, Lee JS. Cellular senescence: a promising strategy for cancer therapy. BMB Rep 2019;52:35-41. [Crossref] [PubMed]
- Sak A, Groneberg M, Stuschke M. DNA-dependent protein kinase: effect on DSB repair, G2/M checkpoint and mode of cell death in NSCLC cell lines. Int J Radiat Biol 2019;95:1205-19. [Crossref] [PubMed]
- Grundy GJ, Parsons JL. Base excision repair and its implications to cancer therapy. Essays Biochem 2020;64:831-43. Erratum in: Essays Biochem 2020;64:845. [Crossref] [PubMed]
- Kong FR, Lv YH, Yao HM, et al. LncRNA PCAT6 promotes occurrence and development of ovarian cancer by inhibiting PTEN. Eur Rev Med Pharmacol Sci 2019;23:8230-8. [Crossref] [PubMed]
- Xin Y, He X, Zhao W, et al. LncRNA PCAT6 increased cholangiocarcinoma cell proliferation and invasion via modulating miR-330-5p. Am J Transl Res 2019;11:6185-95.
- Liu P, Zhao P, Li B, et al. LncRNA PCAT6 Regulated by YY1 Accelerates the Progression of Glioblastoma via miR-513/IGF2BP1. Neurochem Res 2020;45:2894-902. [Crossref] [PubMed]
- Mahmoud AM, Ali MM. Methyl Donor Micronutrients that Modify DNA Methylation and Cancer Outcome. Nutrients 2019;11:608. [Crossref] [PubMed]
- Zafon C, Gil J, Pérez-González B, et al. DNA methylation in thyroid cancer. Endocr Relat Cancer 2019;26:R415-39. [Crossref] [PubMed]
- Hata A, Lieberman J. Dysregulation of microRNA biogenesis and gene silencing in cancer. Sci Signal 2015;8:re3. [Crossref] [PubMed]


