Development and validation of a chromatin regulator signature for predicting prognosis hepatocellular carcinoma patient
Highlight box
Key findings
• Our study presents a chromatin regulator-based prognosis model for Hepatocellular carcinoma (HCC).
What is known and what is new?
• Chromatin regulators involve in cancer development.
• Development and validation of a prognosis model based on chromatin regulators for HCC.
What is the implication, and what should change now?
• These findings offer insights into HCC prognosis and treatment, with implications for personalized medicine and improved patient outcomes. Future studies may explore its application in guiding treatment decisions, improving patient outcomes, and advancing our understanding of HCC pathogenesis.
Introduction
Hepatocellular carcinoma (HCC) is the most prevalent primary hepatic neoplasm, constituting 75–85% of all primary liver tumors and ranking as the fifth most common cancer worldwide. It is also the third leading cause of cancer-related deaths globally with approximately 830,000 deaths reported by WHO in 2020 (1). HCC exhibits notable gender disparities, with men experiencing 2–3 times higher morbidity and mortality rates than women (2). Despite substantial progress in diagnostic and therapeutic modalities, including surgical intervention, transarterial chemoembolization (TACE), targeted therapies, and immunotherapy, the overall survival (OS) rate for advanced HCC patients remains suboptimal (3). Accumulating evidence suggests that multiple gene expression profiles are pivotal in risk stratification and prognostic prediction for cancer patients (4-6). For example, Chen et.al. identified anoikis-related subgroups and prognostic genes in HCC (7). Wang and colleagues latest study suggested that new biomarkers related to disulfidptosis can be used in clinical diagnosis of liver cancer to predict prognosis and treatment targets (8). Ji et al. found that EIF2S2 plays a crucial role in the gene-regulating network of HCC and may be a potential prognostic marker or therapeutic target for HCC patients (9). Luan and colleagues concluded that transcription factor EHF can influence recruitment of neutrophils by mediating the transcription of FGD6 which may contribute to immunotherapy in HCC (10). Lim et al. reported that the dual role of EPHB2 as a cell surface marker and regulator of cancer stemness underscores the importance for future development of specific EPHB2 inhibitors or targeted therapies for potential clinical applications (11). In a related investigation, Chen identified a new cuproptosis-related gene signature that could predict the prognosis of HCC patients (12). Therefore, evaluation of HCC genomics based on specific genes may have significant value for predicting the prognosis and immunotherapy response.
Chromatin regulators (CRs) play a pivotal role in instigating epigenetic changes, considered among the most crucial features of malignancies. Serving as essential regulatory components in epigenetics (13), CRs function as master controllers of gene transcription in normal cells by overseeing histone modifications and chromatin remodeling (14). Their roles in epigenetics classify CRs into 3 primary groups: DNA methylators, histone modifiers, and chromatin remodelers (15). Despite their functional categorization, these groups intricately interrelate in biological processes (BPs). Studies have indicated that aberrant expressions of CRs are associated with various biological functions, including inflammation (16), apoptosis (17), autophagy (18), and proliferation (19). This implies that the deregulation of CRs may contribute to the development of various diseases, including cancer. Abnormal expression of CRs has indeed been established and linked to diverse outcomes in cancer (20). There are various prediction models of other cancer types based on the CR-related genes which can predict prognosis and treatment effect.
Unfortunately, the connection between CRs and HCC has been underexplored in the previous literature. A comprehensive exploration of their roles is essential to advance our understanding of CRs in unraveling the biology of HCC and lay the foundation for future investigations. Our study fills this gap by presenting a comprehensive analysis of the CR signature in HCC and explore its implications in HCC prognosis. Through this, we aimed to illuminate the molecular foundations of HCC, providing new insights for clinical diagnosis and treatment strategies. This study presents a detailed analysis of a CR-based signature designed to predict HCC prognosis and assess its potential clinical relevance. Our investigation encompasses examining immune profiles and the mutational landscape, culminating in developing a CR-related risk score model for HCC. We hypothesize that this model will aid in prognostic assessment and predict responses to both immunotherapy and chemotherapy.
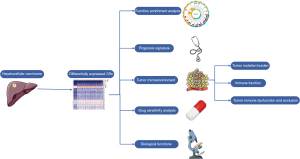
Moreover, we construct an integrated scoring nomogram to refine prognostic stratification, enhancing predictive accuracy for individual patients. Finally, to validate our findings, we conducted immunohistochemistry (IHC) and semiquantitative analysis to assess the expression of MRG-binding protein (MRGBP). This comprehensive approach aims to provide a holistic understanding of CRs in the context of HCC, offering valuable insights for research and potential clinical applications (Figure 1). We present this article in accordance with the TRIPOD reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-996/rc).
Methods
Acquisition of data source and preconditioning
We obtained RNA-seq data and matched clinical data from The Cancer Genome Atlas of liver hepatocellular carcinoma (TCGA-LIHC) through the TCGA website (https://portal.gdc.cancer.gov/). We collected long non-coding RNA (lncRNA) and messenger RNA (mRNA) expression values as well as clinical data from 424 samples, including 374 malignancy samples and 50 normal samples. After removing samples with unknown survival status or time, 376 HCC cases remained with clinical data. Ultimately, we compiled data from 370 samples that included both survival information and mRNA expression. TCGA data was divided into the training and testing groups (1:1 ratio). Using training group, risk score for the survivals of patients with HCC was established, and various analysis were performed based on the risk score. Furthermore, IHC for MRGBP which was the most significant predictor for survival was performed using 180 HCC samples in the independent cohort in order to show the upregulation in HCC. We identified 424 cancer-related genes impacting HCC patients from the FACER database (http://bio-bigdata.hrbmu.edu.cn/FACER/). Subsequently, mRNA expression profiles were standardized using the appropriate R package (The R Foundation for Statistical Computing, Vienna, Austria).
Differential analysis
Differential expression analysis was conducted using the limma package in R software. Genes with |logFC| >1 and false discovery rate (FDR) <0.01 were identified as differentially expressed genes (DEGs). We explored up- and down-regulated genes and identified differentially expressed cancer-related genes (DECRs) in HCC by intersecting them with cancer-related genes.
Screening and construction of a prognostic CR-related model
The expression levels, survival time, and survival status of the aforementioned DECRs were compiled across 370 samples. Using the “caret” R package, the samples were randomly and evenly partitioned into a training group and a testing group. Subsequently, various clinical traits of the 2 groups were examined for significant differences.
First, we performed univariate Cox regression analysis with a significant P value <0.05 to obtain candidate DECRs in the training group. Then, least absolute shrinkage and selection operator (LASSO) regression was performed on these DECRs to further remove the less relevant DECRs, and finally multivariate Cox regression was performed with the selected DECRs to build a risk score model.
The risk score for each sample is calculated as: . Coef indicates the coefficient value, and x indicates the expression level of selected DECRs.
The predictive ability of the prognostic model and validation of the model
By using the “survival” R package, univariate and multivariate independent prognostic analyses were used to plot the model with other clinical traits in forest plots and examine P values to evaluate whether the model could be used as a predictor independent of other clinical traits.
Sample risk scores were ranked from low to high; risk scores, survival, and DECRs expression in the risk model were visualized, and samples were divided into high-risk and low-risk groups using the median. Receiver operating characteristic (ROC) curves at 1, 3, and 5 years were drawn to observe the predictive effect of the model on the prognosis of HCC patients, and concordance index (C-index) curves were drawn to compare the accuracy of this model in predicting the prognosis with other clinical traits.
To evaluate the prognostic value of the model in HCC patients, we analyzed the OS and progression-free survival (PFS) of the high- and low-risk samples by Kaplan-Meier analysis to observe whether there were differences between the high- and low-risk groups.
We integrated the risk formula score with demographic and clinical variables such as age, gender, stage, grade, and tumor-node-metastasis (TNM) stage to formulate a nomogram capable of predicting the 1-, 3-, and 5-year survival probabilities for HCC patients. Subsequently, we validated its accuracy by comparing the actual survival status with the predicted outcomes.
Functional enrichment analyses and gene set enrichment analysis (GSEA)
We performed Gene Ontology (GO) functional enrichment analysis on DECRs. By using the ‘org.Hs.eg.db’ R package, we converted gene names to gene id. Then, we performed enrichment analysis to obtain the gene set enrichment results. The top 6 results in BP, cellular components (CC), and molecular function (MF) were used to draw the GO circle diagram. Statistical significance was determined by P value <0.05 and FDR <0.05 during the process.
DECRs were subjected to Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analysis. Similar to GO functional enrichment analysis, the ‘clusterProfiler’ R package was used to obtain results for gene set enrichment. The most significant 30 pathways were plotted as histograms. Statistical significance was determined by P value <0.05 and FDR <0.25 during the process.
For GSEA, we ranked DECRs in order of high to low expression, combined with channel-related gene sets downloaded from the GSEA database. The 5 most significantly enriched pathways in the high- and low-risk groups were visualized.
Tumor immune microenvironment analysis
Combined with gene expression in various immune cells and each sample, the ‘CIBERSORT’ R package was called to calculate the relative content of immune cells in these samples. Samples with insufficient accuracy were removed at P<0.05, and differential analysis was performed in the remaining samples to observe the differences in the contents of different types of immune cells between high- and low-risk groups. Then ‘GSVA’ and ‘GSEABase’ R packages were called for single sample gene set enrichment analysis (ssGSEA) to observe which immune-related functions were different in the high- and low-risk groups.
Tumor mutation burden (TMB) and tumor immune dysfunction and exclusion (TIDE)
The gene mutation data were stratified into 2 groups based on the risk score model to examine the differences in gene mutations between the high- and low-risk groups. The findings were visualized using the ‘maftools’ R package.
All samples were divided into 2 groups according to the level of TMB for survival analysis to observe whether there was a difference in OS between the 2 groups of samples, and then the samples were divided into 4 groups for survival analysis according to the combined risk score of tumor mutation load to observe whether there was an OS difference.
The expression level of the uploaded gene in each sample was used to obtain the TIDE score of each sample in http://tide.dfci.harvard.edu/, so as to compare whether there was a difference in the potential of immune escape between the high- and low-risk groups. Then, we could predict the efficacy of immunotherapy for samples at different risks.
Drug sensitivity analysis
In addition to immunotherapy, we can observe the difference in the efficacy of traditional chemotherapeutic drugs for HCC between high- and low-risk groups using the ‘oncoPredict’ R package (P<0.001) in order to guide targeted medication for patients of different risks.
Tissue sampling and clinical data
HCC and adjacent tumor tissues (>2 cm) were procured from patients who underwent radical surgical resection of HCC at the Department of Hepatobiliary Pancreatic Splenic Surgery, Affiliated Hospital of Nantong University, Jiangsu Province, China, during the period from January 2013 to June 2020. Fresh tissues were carefully aliquoted and stored at −80 °C for subsequent total protein and RNA extraction. Formalin-fixed tissues were preserved at −4 °C for the construction of tissue microarrays. A rigorous histopathological examination conducted by two experienced pathologists confirmed the identity of all specimens as HCC.
Patient-related data, encompassing age, gender, earliest diagnosed date, survival time, survival status, and TNM staging, were extracted from the hospital’s information system. Exclusion criteria were applied to eliminate patients with Class C Child-Pugh scores, positive surgical margins, preoperative interventions, history of radiotherapy, chemotherapy, targeted therapy, immunotherapy, or incomplete clinical data. Follow-up data were meticulously collected to document postoperative survival.
IHC was performed on 180 pairs of HCC and adjacent tumor tissues (see Table S1). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All samples were acquired with informed consent from the patients, and the study protocol received approval from the Ethics Committee of the Affiliated Hospital of Nantong University (No. 2019-K021).
IHC and semiquantitative analysis
Diluted MRGBP polyclonal antibodies were evenly applied to a tissue chip containing 180 pairs of HCC tissues and adjacent tumor liver tissues. Subsequently, the chip was incubated overnight in a moisture chamber at 4 °C. The corresponding secondary antibodies were uniformly added to the tissue chip, followed by a 30-minute incubation at room temperature. We captured 3 representative images from each sample using a microscope and subjected to double-blind analysis by 2 senior pathologists, both blinded to patient clinical outcomes.
The H-score was determined based on the intensity of nuclear staining and the proportion of labeled tumor cells (21). Briefly, nuclear staining intensity was graded as 0 (no staining), 1 (weak), 2 (moderate), 3 (strong), and employed in the following formula: (% of positive cells, intensity 3 × 3) + (% of positive cells, intensity 2 × 2) + (% of positive cells, intensity 1 × 1) = H-score.
Statistical analysis
All statistical analyses were conducted using R software (version 4.3.0). A significance level of P<0.05 was adopted unless explicitly indicated otherwise. The Wilcoxon test or Student’s t-test was employed for evaluating differences between 2 groups. Kaplan-Meier survival analysis was performed, and the log-rank test was utilized to compare the survival times among different groups.
Results
Establishment of CR-based signature
Through differential expression analysis, we obtained 214 DEGs, of which 3 were down-regulated and 211 were up-regulated in the TCGA-LIHC dataset. The top 50 DECRs with the most significant differences are illustrated in Figure 2A. To assess the prognostic value of CRs, univariate Cox regression analysis was conducted on these deregulated DECRs, identifying 115 of them with prognostic significance (Figure S1). Subsequently, LASSO Cox regression analysis was employed to construct a prognostic signature for HCC patients (Figure 2B,2C). The resulting risk model effectively comprised 3 genes (BMI1, CBX2, and MRGBP), as outlined in Table 1.
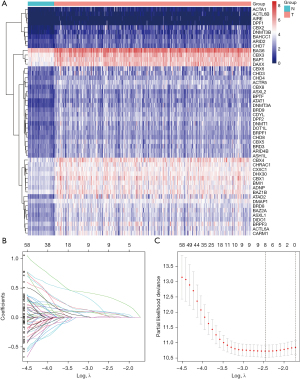
Table 1
| Gene symbol | Full name | Risk coefficient |
|---|---|---|
| BMI1 | B-lymphoma Mo-MLV insertion region 1 | 0.427156094892787 |
| CBX2 | Chromobox homolog 2 | 0.236588727370792 |
| MRGBP | Mortality factor on chromosome 4-related gene-binding protein | 0.674398457243853 |
The calculation of the risk score utilized the pertinent coefficients from the 3 DECRs, following the formula: risk score = (0.427 × BMI1 expression) + (0.237 × CBX2 expression) + (0.674× MRGBP expression).
Differential analysis of diverse clinical traits within the training and testing groups, obtained through equal scoring, revealed P values >0.05 for age, gender, stage, grade, and TNM stage differences between the 2 groups, indicating no significant distinctions (Table 2). Utilizing this grouping, 3 cohorts were employed to evaluate and validate the prognostic value of the model in this study, comprising the training group (N=185), the testing group (N=185), and the overall group (N=370).
Table 2
| Covariates | Total group, n (%) | Testing group, n (%) | Training group, n (%) | P value |
|---|---|---|---|---|
| Age | 0.5909 | |||
| ≤65 years | 232 (62.7) | 119 (64.32) | 113 (61.08) | |
| >65 years | 138 (37.3) | 66 (35.68) | 72 (38.92) | |
| Gender | 0.8246 | |||
| Female | 121 (32.7) | 59 (31.89) | 62 (33.51) | |
| Male | 249 (67.3) | 126 (68.11) | 123 (66.49) | |
| Grade | 0.0771 | |||
| G1 | 55 (14.86) | 23 (12.43) | 32 (17.3) | |
| G2 | 177 (47.84) | 87 (47.03) | 90 (48.65) | |
| G3 | 121 (32.7) | 61 (32.97) | 60 (32.43) | |
| G4 | 12 (3.24) | 10 (5.41) | 2 (1.08) | |
| Unknown | 5 (1.35) | 4 (2.16) | 1 (0.54) | |
| Stage | 0.8315 | |||
| Stage I | 171 (46.22) | 82 (44.32) | 89 (48.11) | |
| Stage II | 85 (22.97) | 45 (24.32) | 40 (21.62) | |
| Stage III | 85 (22.97) | 44 (23.78) | 41 (22.16) | |
| Stage IV | 5 (1.35) | 3 (1.62) | 2 (1.08) | |
| Unknown | 24 (6.49) | 11 (5.95) | 13 (7.03) | |
| T | 0.164 | |||
| T1 | 181 (48.92) | 87 (47.03) | 94 (50.81) | |
| T2 | 93 (25.14) | 51 (27.57) | 42 (22.7) | |
| T3 | 80 (21.62) | 42 (22.7) | 38 (20.54) | |
| T4 | 13 (3.51) | 3 (1.62) | 10 (5.41) | |
| Unknown | 3 (0.81) | 2 (1.08) | 1 (0.54) | |
| M | >0.99 | |||
| M0 | 266 (71.89) | 134 (72.43) | 132 (71.35) | |
| M1 | 4 (1.08) | 2 (1.08) | 2 (1.08) | |
| Unknown | 100 (27.03) | 49 (26.49) | 51 (27.57) | |
| N | >0.99 | |||
| N0 | 252 (68.11) | 128 (69.19) | 124 (67.03) | |
| N1 | 4 (1.08) | 2 (1.08) | 2 (1.08) | |
| Unknown | 114 (30.81) | 55 (29.73) | 59 (31.89) |
TCGA, The Cancer Genome Atlas.
Validation of CR-based signature
The Kaplan-Meier survival curves for all groups are presented in Figure 3A-3F, revealing that patients with a high-risk score tended to exhibit lower survival probabilities and experienced earlier mortality (or metastasis) compared to those with a low-risk score. The area under the curve (AUC) values for the remaining dataset exceeded 0.65, with the exception of the 5-year AUC values, which were 0.604 for the testing group (Figure 3G-3I). These findings indicate that our risk model demonstrates a robust predictive effect on the prognosis of HCC patients.
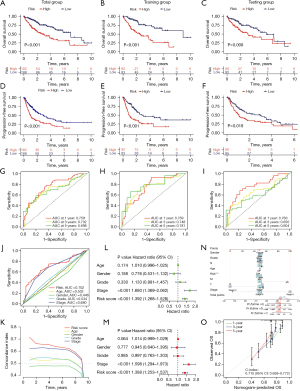
In the 3-year ROC curves, the AUC values for risk scores surpassed those for other clinical traits, suggesting that the utilization of risk scores provides a more accurate prediction of the survival of HCC patients compared to other clinical traits (Figure 3J,3K).
Both univariate and multivariate independent prognostic analyses showed P<0.05, indicating that the model can be used as a predictive tool independent of other clinical traits (Figure 3L,3M).
To predict the 1-, 3-, and 5-year survival probabilities for each sample, we constructed a nomogram containing various clinical traits and risk scores (Figure 3N). A higher score means a lower probability of survival. For example, a patient with HCC who scored 392 had a 1-year survival rate of 0.929, a 3-year survival rate of 0.861, and a 5-year survival rate of 0.81. Calibration images showed that nomograms predicted 1-, 3-, and 5-year OS were in good agreement with actual values and accuracy (Figure 3O).
In summary, this risk model has been validated as a tool to predict prognosis.
Functional enrichment analyses and GSEA
The top 6 with the highest significance among the GO enrichment analysis results were selected and integrated into a circle diagram (Figure 4A). As can be seen in the figure 4A, in BP, these DECRs were mainly involved in these biological functions such as nuclear division, organelle fission, mitotic nuclear division, chromosome segregation, nuclear chromosome segregation, and sister chromatid segregation; in CC, cell components such as chromosomal region, chromosome, centromeric region, condensed chromosome, the CDC45/RecJ, MCM, GINS (CMG) complex, and DNA replication preinitiation complex were involved; in MF, these risk DEGs were mainly involved in MFs such as single-stranded DNA helicase activity, tubulin binding, microtubule binding, DNA helicase activity, ATP-dependent activity acting on DNA, and catalytic activity acting on DNA.

KEGG pathway analysis showed these DECRs were significantly associated with cell cycle pathways, DNA replication, extracellular matrix (ECM)-receptor interaction, and protein digestion and absorption (Figure 4B).
Via GSEA, we noted enrichments in cell cycle, cytokine receptor interaction, ECM receptor interaction, hematopoietic cell lineage, and neuroactive ligand receptor interaction in the high-risk group. Conversely, the low-risk group exhibited enrichments in drug metabolism cytochrome P450, fatty acid metabolism, glycine serine and threonine metabolism, primary bile acid biosynthesis, and retinol metabolism (Figure 4C,4D).
Tumor immune microenvironment analysis
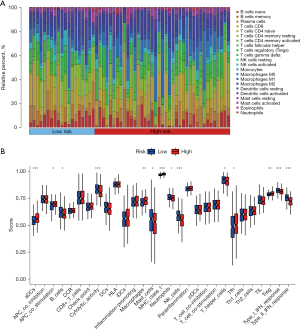
After selecting eligible samples, their relative immune cell content was visualized and analyzed for differences. It could be observed that monocytes were significantly more frequent in low-risk samples than in high-risk samples, whereas macrophages M0 cells were significantly more frequent in high-risk samples than in low-risk samples (Figure 5A). Among various immune-related functions, B cells, cytolytic activity, mast cells, neutrophils, NK cells, T helper cells, type I interferon (IFN) response, and type II IFN response were significantly active in the low-risk group compared with the high-risk group; activated dendritic cells (aDCs), antigen-presenting cell (APC) co stimulation, macrophages, major histocompatibility complex (MHC) class I, T follicular helper (Tfh), and tumor-infiltrating lymphocyte (TIL) was significantly active in the high-risk group compared with the low-risk group (Figure 5B).
TMB and TIDE
TMB analysis of samples from the high- and low-risk groups showed that the gene tp53 had the most mutations in samples from the high-risk group whereas the gene CTNNB1 had the most mutations in samples from the low-risk group. Missense mutations were the most frequent mutation type in most of the mutated genes in both groups (Figure 6A).
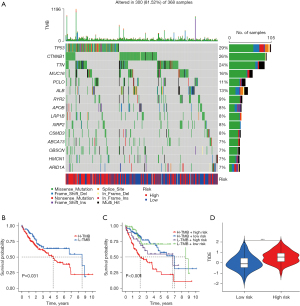
At the same time point, significantly fewer samples survived in the high TMB group than in the low TMB group. In the 4 groups that combined the TMB with the risk model score to divide the samples, it could be observed that the low TMB + low risk group was located at the top of the graph whereas the high TMB + high risk group was located at the bottom of the graph meaning that they had the best and worst prognosis, respectively (Figure 6B,6C).
A notable distinction in TIDE scores was observed between the high- and low-risk groups, with the low-risk group exhibiting lower TIDE scores. This suggests that individuals with low-risk HCC may experience enhanced effectiveness with immunotherapy (Figure 6D).
Drug sensitivity analysis
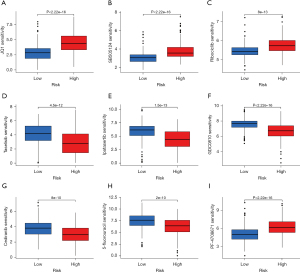
The results showed that PF-4708671, JQ1, ribociclib, and SB505124 could obtain better efficacy in the treatment of high-risk group samples, whereas taselisib, ipatasertib, GDC0810, cediranib, and 5-fluorouracil could obtain better efficacy in the treatment of low-risk group samples. These findings are conducive to the development of accurate treatment plans for high- and low-risk groups, respectively (Figure 7).
MRGBP is highly expressed and predicts poor prognosis in HCC
When considering all signature genes, MRGBP emerged as the most significant predictor of survival in multivariate Cox regression analysis due to its expression level and risk coefficient. It is located in nucleoplasm. Subsequently, we delved deeper into the mRNA expression levels and prognostic implications of MRGBP, revealing its upregulation in HCC tissues compared to normal tissues and its association with a shorter OS (Figure 8A,8B). IHC was employed to assess MRGBP expression in tissue chips, encompassing 180 pairs of HCC tissues and their corresponding tumor-adjacent liver tissues. Noticeably, the expression of MRGBP exhibited marked differences between HCC and tumor-adjacent liver tissues (Figure 8C). Across the 180 pairs of tissue sections in our study, the disparities in MRGBP expression in HCC tissues and tumor-adjacent liver tissues were statistically significant, further indicating an association with poor prognosis in HCC (Figure 8D).

Discussion
The discovery of novel risk factors plays a crucial role in enhancing the diagnosis and prognosis of HCC, aiding healthcare professionals in assessing patient risk and tailoring personalized treatment strategies. In recent times, there has been a continuous evolution in the exploration of diagnostic and prognostic markers for HCC, paralleling the progress in information technology. For example, Shen et al. revealed that recognizing MITD1 as a novel biomarker for HCC could offer insights into how alterations in cytokinesis and the immune milieu contribute to the development of liver cancer (22). Upon more in-depth analysis, MITD1 could be a prognostic indicator for human HCC. Subsequently, Xiang et al. identified 3 pivotal genes (CASKIN1, EMR3, and GBP5) through screening mRNA-seq sequencing data and establishing a stem cell index derived from TCGA-LIHC mRNA profiles (23).
Nonetheless, dependable clinical diagnostic and prognostic biomarkers are required to be substantiated by data for HCC. Despite numerous studies illustrating the diverse roles of CRs in tumor progression (24-26), only a limited number have undertaken thorough examinations of their clinical relevance in HCC. As integral components of the epigenetic machinery, CRs regulate the transcriptional process of substantial cell genes, including oncogenes. Consequently, alterations in their activity wield profound influence over the overarching landscape of genetic expression and the intricate signaling networks that underpin cellular health. This regulatory paradigm significantly potentiates the proliferative capabilities of oncogenes, thereby laying the groundwork for the eventual onset of oncogenesis. Therefore, it is essential to conduct studies targeting HCC based on CRs.
In this study, we initially explored the TCGA database to identify CRs that exhibited differential expression in 214 HCC tissues compared to normal liver tissues. Subsequently, we developed a CR-based signature by conducting a thorough investigation of the biological pathways associated with these 214 DECRs. These DECRs were meticulously selected based on their biological significance, leading to the creation of a risk model centered around 3 key genes: BMI1, CBX2, and MRGBP. Our risk model demonstrated promising predictive capabilities for HCC patient prognosis, as evidenced by Kaplan-Meier survival curves and ROC curve analysis. Consistently, the results indicated that patients with higher risk scores experienced poorer survival outcomes, highlighting the potential utility of this signature as a prognostic tool. Importantly, the risk model’s effectiveness was further affirmed by its independence from other clinical traits, as demonstrated in both univariate and multivariate independent prognostic analyses.
Furthermore, we constructed a nomogram to predict 1-, 3-, and 5-year survival probabilities for individual patients, providing a practical tool for clinicians to assess and communicate prognosis. Additionally, our GSEA unveiled distinct molecular pathways enriched in high- and low-risk groups, offering valuable insights into potential therapeutic targets and mechanisms underlying HCC progression.
Tumor immune microenvironment analysis revealed intriguing disparities in immune cell composition and activity between high- and low-risk groups. Notably, monocytes were more prevalent in low-risk samples, whereas macrophages M0 were more abundant in high-risk samples. These findings underscore the complex interplay between the CR-based signature and the immune response in HCC.
TMB analysis suggested a correlation between genetic mutations and risk groups, with specific genes displaying varying mutation patterns. Significantly, the combined assessment of TMB and the risk model allowed for the identification of distinct patient subgroups with markedly different survival outcomes. Furthermore, the lower TIDE scores in the low-risk group suggest the potential for enhanced immunotherapy efficacy in this subgroup.
Lastly, our drug sensitivity analysis identified specific chemotherapeutic agents that may be more effective in high- or low-risk groups, thereby offering personalized treatment options based on the CR-based signature.
Our risk model comprised 3 genes: BMI1, CBX2, and MRGBP. These genes have been repeatedly implicated in prior studies for their roles in tumorigenesis. BMI1 is a recognized proto-oncogene that contributes to the initiation and progression of various malignancies. In the context of HCC, BMI1 exhibits upregulated expression. It influences HCC development through diverse mechanisms, including its impact on the INK4a/ARF locus, involvement in the NF-κB signaling pathway, and modulation of the PTEN/PI3K/AKT signaling pathway (27-31). Moreover, BMI1 expression has been found to be closely associated with both HCC prognosis and recurrence (32). CBX2, a member of the chromobox family of proteins, is a pivotal component of the polycomb group complex. Previous investigations have revealed the involvement of CBX2 in the development and progression of several cancers, such as breast cancer (33), lung adenocarcinoma (34), and gastric cancer (35). Notably, a study conducted by Mao and Tian have shown that the knockdown of CBX2 expression in HCC cells resulted in increased HCC cell apoptosis, suppressed HCC cell proliferation, and enhanced YAP phosphorylation, both in vitro and in vivo (36). Xu found that CBX2-mediated suppression of SIAH2 triggers WNK1 accumulations to promote glycolysis in HCC (37). This suggests that CBX2 holds potential as a therapeutic target for HCC treatment. MRGBP is a transcription factor with widespread involvement in various physiological and pathological processes. Multiple studies have explored the relationship between MRGBP expression levels and the prognosis of various malignant tumors. MRGBP amplification is frequently observed in numerous cancer types, including lung (38), head and neck squamous cell carcinoma (39), prostate (40), and pancreatic cancers (41). Huang’s research revealed a correlation between elevated MRGBP expression, cancer advancement, diminished survival rates, and heightened levels of immune infiltration in HCC. This indicates that MRGBP could be a novel prognostic biomarker associated with immune infiltrates (42). In our investigation, IHC experiments confirmed that MRGBP stimulates the malignant progression of HCC. These results align with the findings from bioinformatic analyses and Huang’s study.
This comprehensive investigation provides valuable insights into the clinical significance of CRs in HCC and underscores the potential for personalized therapeutic approaches and improved patient outcomes.
Furthermore, to gain a comprehensive understanding of the mechanisms through which CRs influence the biological behavior of HCC cells, additional experimental validation is crucial. Future studies should aim to elucidate the specific pathways and molecular interactions involved, building upon the foundation laid by this investigation. Moreover, to enhance the generalizability of our prognostic model, it is imperative to subject it to rigorous validation in multicenter clinical cohorts. This will not only bolster the reliability of our findings but also contribute to the broader applicability of the CR-based signature in diverse clinical settings.
Conclusions
In conclusion, our study presents a robust CR-based signature that holds promise as a valuable prognostic tool for HCC patients. The findings from this study lay the foundation for further research into the clinical utility and biological mechanisms of the CR-based signature. It is recommended that future studies delve into the practical implementation of this signature in clinical settings, exploring its impact on treatment strategies and patient management. Additionally, a more in-depth exploration of the molecular pathways influenced by the CR-based signature could unveil novel therapeutic targets. Such endeavors will not only refine the prognostic significance of the signature but also contribute to the ongoing evolution of personalized medicine in the context of HCC. In summary, this study enriches our understanding of the multifaceted role of chromatin regulation in HCC, emphasizing its potential clinical implications.
Acknowledgments
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-996/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-996/dss
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-996/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-996/coif). R.I. receives payment or honoraria for lectures from Cook and Boston Scientific, outside the submitted work. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All samples were acquired with informed consent from the patients, and the study protocol received approval from the Ethics Committee of the Affiliated Hospital of Nantong University (No. 2019-K021).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Ruggieri A, Barbati C, Malorni W. Cellular and molecular mechanisms involved in hepatocellular carcinoma gender disparity. Int J Cancer 2010;127:499-504. [Crossref] [PubMed]
- Chen Z, Xie H, Hu M, et al. Recent progress in treatment of hepatocellular carcinoma. Am J Cancer Res 2020;10:2993-3036.
- Tang Y, Guo C, Yang Z, et al. Identification of a Tumor Immunological Phenotype-Related Gene Signature for Predicting Prognosis, Immunotherapy Efficacy, and Drug Candidates in Hepatocellular Carcinoma. Front Immunol 2022;13:862527. [Crossref] [PubMed]
- Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9:34. [Crossref] [PubMed]
- Roessler S, Long EL, Budhu A, et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology 2012;142:957-66.e12. [Crossref] [PubMed]
- Chen Y, Huang W, Ouyang J, et al. Identification of Anoikis-Related Subgroups and Prognosis Model in Liver Hepatocellular Carcinoma. Int J Mol Sci 2023;24:2862. [Crossref] [PubMed]
- Wang T, Guo K, Zhang D, et al. Disulfidptosis classification of hepatocellular carcinoma reveals correlation with clinical prognosis and immune profile. Int Immunopharmacol 2023;120:110368. [Crossref] [PubMed]
- Ji P, Wang H, Cheng Y, et al. Prognostic prediction and gene regulation network of EIF2S2 in hepatocellular carcinoma based on data mining. J Gastrointest Oncol 2021;12:3061-78. [Crossref] [PubMed]
- Luan M, Tian X, Zhang D, et al. Identifying the potential regulators of neutrophils recruitment in hepatocellular carcinoma using bioinformatics method. Transl Cancer Res 2021;10:724-37. [Crossref] [PubMed]
- Lim JJ, Chow EKH, Toh TB. Eph receptor B2 (EPHB2) regulates cancer stem cell-like properties in hepatocellular carcinoma. Stem Cell Investig 2022;9:5. [Crossref] [PubMed]
- Chen Y, Tang L, Huang W, et al. Identification of a prognostic cuproptosis-related signature in hepatocellular carcinoma. Biol Direct 2023;18:4. [Crossref] [PubMed]
- Lu J, Xu J, Li J, et al. FACER: comprehensive molecular and functional characterization of epigenetic chromatin regulators. Nucleic Acids Res 2018;46:10019-33. [Crossref] [PubMed]
- Shu XS, Li L, Tao Q. Chromatin regulators with tumor suppressor properties and their alterations in human cancers. Epigenomics 2012;4:537-49. [Crossref] [PubMed]
- Plass C, Pfister SM, Lindroth AM, et al. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat Rev Genet 2013;14:765-80. [Crossref] [PubMed]
- Marazzi I, Greenbaum BD, Low DHP, et al. Chromatin dependencies in cancer and inflammation. Nat Rev Mol Cell Biol 2018;19:245-61. [Crossref] [PubMed]
- Li T, Yang J, Yang B, et al. Ketamine Inhibits Ovarian Cancer Cell Growth by Regulating the lncRNA-PVT1/ EZH2/p57 Axis. Front Genet 2020;11:597467. [Crossref] [PubMed]
- Chu Y, Chen W, Peng W, et al. Amnion-Derived Mesenchymal Stem Cell Exosomes-Mediated Autophagy Promotes the Survival of Trophoblasts Under Hypoxia Through mTOR Pathway by the Downregulation of EZH2. Front Cell Dev Biol 2020;8:545852. [Crossref] [PubMed]
- Chen J, Wang F, Xu H, et al. Long Non-Coding RNA SNHG1 Regulates the Wnt/β-Catenin and PI3K/ AKT/mTOR Signaling Pathways via EZH2 to Affect the Proliferation, Apoptosis, and Autophagy of Prostate Cancer Cell. Front Oncol 2020;10:552907. [Crossref] [PubMed]
- Yan XJ, Xu J, Gu ZH, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet 2011;43:309-15. [Crossref] [PubMed]
- Detre S, Saclani Jotti G, Dowsett M A. "quickscore" method for immunohistochemical semiquantitation: validation for oestrogen receptor in breast carcinomas. J Clin Pathol 1995;48:876-8. [Crossref] [PubMed]
- Shen H, Wang Z, Ren S, et al. Prognostic biomarker MITD1 and its correlation with immune infiltrates in hepatocellular carcinoma (HCC). Int Immunopharmacol 2020;81:106222. [Crossref] [PubMed]
- Xiang S, Li J, Shen J, et al. Identification of Prognostic Genes in the Tumor Microenvironment of Hepatocellular Carcinoma. Front Immunol 2021;12:653836. [Crossref] [PubMed]
- Li X, Huo X, Zhao C, et al. A novel chromatin regulator signature predicts the prognosis, clinical features and immunotherapy of colon cancer. Epigenomics 2022;14:1325-41. [Crossref] [PubMed]
- Liao WB, Liu L. Identification of a chromatin regulator signature for predicting prognosis of prostate cancer patient. Eur Rev Med Pharmacol Sci 2023;27:275-90. [Crossref] [PubMed]
- Bélanger S, Haupt S, Faliti CE, et al. The Chromatin Regulator Mll1 Supports T Follicular Helper Cell Differentiation by Controlling Expression of Bcl6, LEF-1, and TCF-1. J Immunol 2023;210:1752-60. [Crossref] [PubMed]
- Wang R, Fan H, Sun M, et al. Roles of BMI1 in the Initiation, Progression, and Treatment of Hepatocellular Carcinoma. Technol Cancer Res Treat 2022;21:15330338211070689. [Crossref] [PubMed]
- Chen MH, Fu LS, Zhang F, et al. LncAY controls BMI1 expression and activates BMI1/Wnt/β-catenin signaling axis in hepatocellular carcinoma. Life Sci 2021;280:119748. [Crossref] [PubMed]
- Li B, Chen Y, Wang F, et al. Bmi1 drives hepatocarcinogenesis by repressing the TGFβ2/SMAD signalling axis. Oncogene 2020;39:1063-79. [Crossref] [PubMed]
- Ma DQ, Zhang YH, Ding DP, et al. Effect of Bmi-1-mediated NF-κB signaling pathway on the stem-like properties of CD133+ human liver cancer cells. Cancer Biomark 2018;22:575-85. [Crossref] [PubMed]
- Jacobs JJ, Kieboom K, Marino S, et al. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 1999;397:164-8. [Crossref] [PubMed]
- Ruan ZP, Xu R, Lv Y, et al. Bmi1 knockdown inhibits hepatocarcinogenesis. Int J Oncol 2013;42:261-8. [Crossref] [PubMed]
- Chen WY, Zhang XY, Liu T, et al. Chromobox homolog 2 protein: A novel biomarker for predicting prognosis and Taxol sensitivity in patients with breast cancer. Oncol Lett 2017;13:1149-56. [Crossref] [PubMed]
- Hu FF, Chen H, Duan Y, et al. CBX2 and EZH2 cooperatively promote the growth and metastasis of lung adenocarcinoma. Mol Ther Nucleic Acids 2022;27:670-84. [Crossref] [PubMed]
- Zeng M, Li B, Yang L, et al. CBX2 depletion inhibits the proliferation, invasion and migration of gastric cancer cells by inactivating the YAP/β-catenin pathway. Mol Med Rep 2021;23:137. [Crossref] [PubMed]
- Mao J, Tian Y, Wang C, et al. CBX2 Regulates Proliferation and Apoptosis via the Phosphorylation of YAP in Hepatocellular Carcinoma. J Cancer 2019;10:2706-19. [Crossref] [PubMed]
- Xu Z, Wu Y, Yang M, et al. CBX2-mediated suppression of SIAH2 triggers WNK1 accumulations to promote glycolysis in hepatocellular carcinoma. Exp Cell Res 2023;426:113513. [Crossref] [PubMed]
- Dai J, Li Z, Amos CI, et al. Systematic analyses of regulatory variants in DNase I hypersensitive sites identified two novel lung cancer susceptibility loci. Carcinogenesis 2019;40:432-40. [Crossref] [PubMed]
- Zhao C, Wei C, Chen X, et al. MRGBP: A New Factor for Diagnosis and Prediction of Head and Neck Squamous Cell Carcinoma. Biomed Res Int 2022;2022:7281120. [Crossref] [PubMed]
- Ito S, Ueda T, Ueno A, et al. A genetic screen in Drosophila for regulators of human prostate cancer progression. Biochem Biophys Res Commun 2014;451:548-55. [Crossref] [PubMed]
- Ding F, Zhang S, Gao S, et al. MiR-137 functions as a tumor suppressor in pancreatic cancer by targeting MRGBP. J Cell Biochem 2018;119:4799-807. [Crossref] [PubMed]
- Huang J, Chen X, Zhu W. MRGBP is a potential novel prognostic biomarker and is correlated with immune infiltrates in hepatocellular carcinoma. Medicine (Baltimore) 2021;100:e25234. [Crossref] [PubMed]