
Minimal residual disease monitoring via ctDNA: a case report of Lynch syndrome with synchronous colorectal cancer and review of literature
Highlight box
Key findings
• This case report underscores the complex nature of minimal residual disease (MRD) monitoring using circulating tumor DNA (ctDNA) in synchronous colorectal cancers (CRC). The unique case highlights distinct genomic alterations in each tumor, emphasizing the challenges of tumor informed ctDNA assays as a surrogate for MRD. This holds particular importance in the context of concurrent malignancies, adding intricacy to ctDNA’s function as an MRD indicator in localized CRC.
What is known and what is new?
• The prevalence of synchronous CRC has been reported to be around 3%, rising to 10% or more in patients with Lynch syndrome specifically. Tumor-informed assays, while advantageous for their high specificity, may result in false negatives, as evidenced by our patient’s case of synchronous malignancies.
• This study introduces ctDNA’s role in Lynch syndrome-associated synchronous CRC, emphasizing the current limitations of tumor-informed assays in handling genomic heterogeneity as part of surveillance for MRD.
What is the implication, and what should change now?
• The case underscores the evolving role of ctDNA in managing synchronous CRC and Lynch syndrome. Clinicians should incorporate ctDNA assays tailored to each of the individual tumor profiles to achieve accurate monitoring and early detection of recurrence. Future guidelines should address the utility of ctDNA in Lynch syndrome and synchronous CRC, informing surveillance protocols. This emphasizes the urgent need for evidence-based approaches integrating ctDNA to improve outcomes for these high-risk patient populations.
Introduction
The presence of circulating tumor DNA (ctDNA) refers to fragments of DNA from cancerous cells that have entered the bloodstream. The utility of ctDNA as a clinical tool first gained interest approximately two decades ago as a noninvasive method of disease monitoring and genomic analysis (1). Furthermore, the ability to rapidly obtain genomic information without the need for additional radiation exposure or invasive tissue sampling has made ctDNA a promising alternative to traditional tissue genomic assays (2).
While the initial application of ctDNA has been investigated in patients with advanced disease to identify predictive biomarkers for targeted therapy, the potential role for ctDNA has been expanded to cancer patients with early-stage disease including the assessment of minimal residual disease (MRD) (3,4). MRD is defined as the presence of residual malignant cells after curative treatment which share phenotypic and genetic similarities to the original tumor (5). While there has been interest regarding integration of MRD for surveillance for patients with early-stage cancers as a predictive biomarker, there is a lack of randomized data to support its use (6).
Despite its limitations, ctDNA has been shown to be a reliable indicator of MRD in localized colorectal cancer (CRC) after curative-intent surgery with very high specificity (7). Furthermore, the presence of detectable ctDNA in the post-operative setting has been shown to increase the risk of relapse by 40 times and precedes detected radiographic changes by a median of 4 months (8,9). Given the potential utility and application of MRD in the surveillance of patients with early-stage CRC, guidelines from the National Cancer Institute Colon and Rectal-Anal Task Force regarding the benefits of ctDNA in CRC were published in a Nature Reviews Clinical Oncology article in 2020 (10). Among these guidelines is the diagnostic utility of next generation sequencing (NGS) multigene assays after adjuvant therapy or post-operatively as a surrogate for MRD.
Lynch syndrome, previously known as hereditary nonpolyposis colorectal cancer (HNPCC), is a unique area of research due to various gene mutations associated with DNA mismatch repair (MMR) and the molecular phenotype of high frequency microsatellite instability (MSI-H) malignancies (11). Several genes of the MMR system have been implicated in the development of Lynch syndrome, including MLH1, MSH2, MSH6, and PMS2 (12). Areas within the genome containing repetitive sequences, known as short-tandem repeats or microsatellites, are especially prone to errors during DNA replication in the setting of mismatch repair deficiency (dMMR). Although Lynch syndrome accounts for only 2–4% of CRC, approximately 15% of all CRC tumors have MSI-H (13).
Immune checkpoint inhibitors (ICIs) such as pembrolizumab and nivolumab with or without ipilimumab have changed the treatment paradigm in patients with MSI-H malignancies. The use of ICI directed therapy for MSI-H cancers was first demonstrated in a 2015 phase 2 trial which showed improved outcomes with pembrolizumab in chemorefractory dMMR metastatic CRC and non-CRC cancers (14). The phase 3 KEYNOTE-177 trial showed that treatment with pembrolizumab resulted in superior progression-free survival (PFS) compared to traditional 5-FU based chemotherapy in the first-line setting for advanced MSI-H/dMMR CRC (15). These trials established the role of ICIs in metastatic MSI-H CRC and spares patients from toxicities associated with chemotherapy.
Early-stage MSI-H tumors have been shown in multiple studies to confer a prognostic advantage compared to microsatellite stable (MSS) lesions. A systematic review and meta-analysis of stage II CRC estimated the risk of relapse to be 40% less [hazard ratio (HR) 0.59] with MSI-H compared MSI-L/MSS stage II CRC (16). The impact of MSI status as a prognostic indicator for late-stage CRC is mixed and remains a topic of debate. A meta-analysis of stage III/IV CRC found no difference in disease-free survival (DFS), disease-specific survival (DSS), or overall survival (OS) for MSI-H vs. MSS stage III CRC, and benefit only DFS for MSI-H vs. MSS stage IV CRC (17). As such, detection of MRD has become increasingly utilized to monitor for disease recurrence and gauge effectiveness of adjuvant therapy. The case presented herein highlights the strengths and limitations of ctDNA in a patient harboring two synchronous primary malignancies in the setting of Lynch syndrome. We present this case in accordance with the CARE reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-81/rc).
Case description
We present a case of a 75-year-old male with Lynch syndrome secondary to a germline mutation in MSH6, found to have synchronous rectal and left-sided colon adenocarcinomas. Medical history was significant for squamous cell carcinoma of the vocal cords in 2020 treated with resection and radiation, prostate cancer in 2017 treated with radical prostatectomy, and a large right sided precancerous polyp in 2016 requiring a laparoscopic right hemicolectomy. In Spring 2021, the patient began experiencing tenesmus with rectal bleeding. Diagnostic colonoscopy demonstrated an ulcerated colonic mass 35 cm from the anal verge. Pathology showed moderately differentiated adenocarcinoma invasive to the lamina propria. Immunohistochemical stains of the colonic mass showed loss of nuclear expression of MSH6 and intact expression of MLH1, PMS2, and MSH2. Baseline CEA levels were mildly elevated at 3.3. Germline testing confirmed hereditary mutation in MSH6.
The patient underwent a laparoscopic proctocolectomy with ileostomy. Pathologic findings demonstrated two distinct primary malignancies. A 4.7 cm moderately differentiated adenocarcinoma with invasion through muscularis propria involving pericolorectal tissue was found in the distal transverse colon. A separate 3 cm poorly differentiated mucinous adenocarcinoma with invasion through the muscularis propria/internal sphincter muscle involving mesorectal tissue was found at the distal rectum. A total of 17 out of 39 regional lymph nodes were positive for malignancy. Both primary lesions demonstrated high tumor mutational burden (TMB). There were 74 mut/Mb identified in the transverse colon primary, and 68 mut/Mb identified in the rectal primary, as well as MSI-H in both. The final American Joint Committee on Cancer (AJCC) pathologic staging of each cancer was classified as pT3 and collectively as pT3N2bM0 (stage IIIC).
The patient declined adjuvant chemotherapy in favor of active surveillance. In addition to routine imaging and tracking tumor markers levels, a tumor informed assay calibrated to the patient’s rectal cancer was utilized to monitor ctDNA for MRD. The Natera™ Signatera ™ Residual disease test (MRD) tumor informed assay was sent as part of the active surveillance plan. In this assay, tumor-specific mutation signatures previously detected via whole exome sequencing are identified in peripheral blood samples. Signatera was sent from the rectal tumor primary cancer due to logistical testing limitations which did not allow for two primary tumors to be sent. At the time of publication, the manufacturer states the MRD assay is designed for a single tumor and that multiple simultaneous assays cannot be developed for the same patient. While a different diagnostic assay to monitor MRD, such as a non-tumor informed assay (Guardant Reveal) would have been an alternative consideration, the lessened sensitivity and higher false negative rate led to the decision to use Signatera.
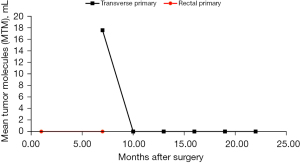
Initial MRD assay levels were undetected approximately 1 month following proctocolectomy. Unfortunately, a 6-month post-operative CT abdomen/pelvis showed worsening retroperitoneal and pelvic lymphadenopathy, concerning for disease recurrence. CEA had increased to 15.4. MRD assay remained negative. Upon request, Signatera MRD assay was sent from the distal transverse colon primary tumor which was positive with an elevated level of 17.58 mean tumor molecules per mL (MTM/mL). Biopsy of the retroperitoneal lymph node confirmed recurrent adenocarcinoma. Genomic profiling of the two primary lesions showed two distinct mutation profiles (Table 1).
Table 1
| Synchronous primary tumors | Genes associated |
|---|---|
| Transverse colon cancer primary | ACVR2A, APC, B2M, CDK12, CDKN2A, FANCE, FBXW7, KEAP1, KMT2B, KMT2C*, MAP3K1, MSH3, PIK3CA, PIK3R1, RASA1,SMARCA4, SOX9, TCF7L2, TSC2 |
| Rectal cancer primary | ARID1A, ARID2, ASXL1, ATM, EP300, ERBB2, ERBB3, GNAS, HRAS, RNF43, GLYR1, KMT2C*, ZBTB7A |
*, conserved mutations between the two primary malignancies.
The patient was started on pembrolizumab for recurrent MSI-H CRC following the results of the MRD assay and CT findings. He was started at a dose of 200 mg every 3 weeks and transitioned to 400 mg every 6 weeks after his 5th cycle. After the 7th cycle, repeat CT abdomen/pelvis showed small volume retroperitoneal and common iliac nodes similar in size to prior imaging but with reduced density, suggesting effect of immunotherapy. No new metastatic disease in the abdomen, pelvis, or chest. Furthermore, CEA remained within normal limits, and MRD testing showed no evidence of ctDNA after initiation of pembrolizumab. Specifically, ctDNA levels were undetected after the 4th cycle of pembrolizumab and have remained undetected 6 months after initiating immunotherapy. Graphic trend of ctDNA monitoring after the proctocolectomy and throughout the treatment course is depicted in Figure 1. To date, the patient has received 9 cycles of pembrolizumab without adverse events and continues with routine imaging and periodic ctDNA levels.
All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Helsinki Declaration (as revised in 2013). Written informed consent for publication of this case report and accompanying images was not obtained from the patient or the relatives after all possible attempts were made.
Discussion
This case highlights multiple aspects of how management and treatment of gastrointestinal malignancies, specifically MSI-H CRC, has shifted over recent years. Improvements in clinical tools including NGS have had profound impacts in outcomes with gastrointestinal malignancies. The case presented demonstrates unique challenges to the field. One of the most distinctive elements of the case was the finding of two simultaneous primary colorectal malignancies. Lynch syndrome is associated with multiple colonic and extracolonic malignancies with an approximate 35–55% risk of developing CRC (18,19). Prior studies have reported a risk of metachronous CRC of approximately 25–35% in Lynch syndrome, with MLH1 and MSH2 mutations conferring higher risk compared to MSH6 and PMS2 (20,21). However, the incidence of synchronous CRC in Lynch syndrome patients is not well established.
A literature review from three pooled population studies estimated a 3.5% prevalence of synchronous primary malignancies occurring for all types of CRC (22). Analysis showed that while patients with synchronous CRC are associated with MSI-H status, many result from sporadic germline MMR mutations. The proposed mechanism is felt to occur via MLH1 promoter hypermethylation and is highly correlated with V600E BRAF mutation (23). Various retrospective studies in patients with synchronous CRC show a prevalence of MSI-H status near 25–35% (24,25). The prevalence of sporadic vs germline MMR mutations in this population remains unclear, although some studies report Lynch syndrome may be more likely to develop synchronous disease (26-28).
While ctDNA has garnered interest as a surrogate biomarker for surveillance with MRD in early-stage CRC, our report highlights potential limitations specifically regarding the finding of concurrent malignancies with unique genetic profiles. Tumor-informed assays utilize multiplex PCR to amplify clonal variants identified via NGS from primary tumor samples. Tumor informed assays have the advantage of high specificity as the test is tailored toward the unique mutations identified from the original tumor sample (7). However, this may lead to false negatives in the case of synchronous malignancies, as occurred for our patient. The radiographic recurrence and subsequent biopsy did not coincide with the 6-month Signatera MRD assay calibrated to the initial rectal primary tumor. Presumably, recurrence in the retroperitoneal lymph node resulted from locoregional spread from the initial transverse colon primary, as the corresponding MRD assay shortly after confirmed biochemical recurrence.
Tumor-specific assays utilize various algorithms to determine the presence of ctDNA in a sample based on prior sequencing data. These algorithms are designed to distinguish true tumor-derived mutations from sequencing errors, clonal hematopoiesis, and other background noise (29). Currently, the Signatera MRD assay is considered positive if at least two tumor-specific variants are detected above threshold (30). Due to the nature of tumor-informed MRD assays targeted towards a single tumor, both synchronous and metachronous malignancies can go undetected. Considering the reported prevalence of synchronous CRC to be approximately 30%, the potential for false negatives is significant and should be factored into clinical decision-making.
Non-tumor informed (also referred as tumor agnostic or plasma only) assays have the advantage of monitoring ctDNA without prior knowledge of molecular alterations, however this approach may lead to false positives (31). These concerns were addressed in a retrospective study of 252 plasma samples from 103 patients with CRC comparing plasma only ctDNA assays against tumor informed assays (7). Results showed comparable sensitivity and specificity between the two types of assays and highlights the potential role for plasma only ctDNA-guided MRD detection. Future application of multiple simultaneous tumor-informed ctDNA assays in synchronous malignancies would also decrease the propensity for false negatives. Alternatively, selecting driver mutations from each malignancy as part of a combined assay may improve sensitivity, but may sacrifice the specificity.
The genomic heterogeneity identified in the patient described herein has also been reported in a review of 23 patients with a combined 50 synchronous primary tumors (32). The study showed that 20 out of 23 cases (87%) expressed unique intra and intertumoral molecular profiles. Interestingly, there was marked variability in clonal variants within individual tumor samples including mutations to KRAS, APC, TP53, PIK3CA, and TGFBR2. As a result of the distinct molecular aberrations associated with synchronous CRCs, there are a multitude of factors to consider including overall prognosis, response to therapy, as well as the diagnostic and ethical implications for ctDNA as a surrogate for MRD.
There are several key studies investigating the clinical utility of ctDNA for MRD in early-stage CRC including the COBRA, CIRCULATE, CIRCULATE-PRODIGE 70, and DYNAMIC-III trials (33-36). The COBRA trial is ongoing as part of a two-phase study of 1,408 patients with resected stage II CRC split into standard-of-care (arm A) and prospective testing with ctDNA (arm B). Patients in arm B with detected levels of ctDNA undergo 6 months of adjuvant FOLFOX chemotherapy with primary endpoint of ctDNA clearance in phase 2 and recurrence-free survival (RFS) in phase 3 (33). The CIRCULATE and CIRCULATE-PRODIGE 70 trials are similar based on selection of resected stage II CRC patients with positive ctDNA randomized to receive adjuvant capecitabine-based or modified FOLFOX chemotherapy, respectively, compared to observation (34,36). Primary endpoints for each of these trials are differences in DFS.
Conclusions
In summary, we present a patient with synchronous CRCs in the setting of Lynch syndrome with confirmed germline mutation in MSH6. Genetic analysis from the primary tumors revealed distinct genomic alterations, adding further complexity to ctDNA tumor informed assays as part of monitoring MRD. Despite the unique molecular characteristics of the two malignancies, the patient had clearance of ctDNA after just four cycles of pembrolizumab. Immunotherapy has resulted in profound impacts in the outcomes of patients with advanced malignancies, particularly for those harboring mutations amenable to targeted treatments such as MSI-H and dMMR CRC. The importance of ctDNA as a surrogate for MRD has been extensively studied as evidenced by the multiple clinical trials discussed previously. The case described herein emphasizes the limitations of tumor informed assays. Tumor-informed assays are currently restricted in their ability to identify a single tumor, potentially overlooking synchronous and metachronous malignancies. Tests designed for multiple tumors or employing tumor-agnostic approaches ought to be considered in clinical decisions for patients with similar conditions. However, there are multiple ongoing investigations which aim to explore future applications of ctDNA and outcomes affecting patient prognosis, treatment, and OS.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the CARE reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-81/rc
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-81/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-81/coif). D.H.A. reports that he receives consulting fees from Exelixis, Genentech, Eisai, Advanced Accelerator Applications, Daiichi Sankyo, and holds stock or stock options in Natera. The other authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017;17:223-38. [Crossref] [PubMed]
- Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem 2015;61:112-23. [Crossref] [PubMed]
- Tie J, Cohen JD, Lahouel K, et al. Circulating Tumor DNA Analysis Guiding Adjuvant Therapy in Stage II Colon Cancer. N Engl J Med 2022;386:2261-72. [Crossref] [PubMed]
- Moding EJ, Nabet BY, Alizadeh AA, et al. Detecting Liquid Remnants of Solid Tumors: Circulating Tumor DNA Minimal Residual Disease. Cancer Discov 2021;11:2968-86. [Crossref] [PubMed]
- Luskin MR, Murakami MA, Manalis SR, et al. Targeting minimal residual disease: a path to cure? Nat Rev Cancer 2018;18:255-63. [Crossref] [PubMed]
- Merker JD, Oxnard GR, Compton C, et al. Circulating Tumor DNA Analysis in Patients With Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol 2018;36:1631-41. [Crossref] [PubMed]
- Parikh AR, Van Seventer EE, Siravegna G, et al. Minimal Residual Disease Detection using a Plasma-only Circulating Tumor DNA Assay in Patients with Colorectal Cancer. Clin Cancer Res 2021;27:5586-94. [Crossref] [PubMed]
- Reinert T, Henriksen TV, Christensen E, et al. Analysis of Plasma Cell-Free DNA by Ultradeep Sequencing in Patients With Stages I to III Colorectal Cancer. JAMA Oncol 2019;5:1124-31. [Crossref] [PubMed]
- Wang Y, Li L, Cohen JD, et al. Prognostic Potential of Circulating Tumor DNA Measurement in Postoperative Surveillance of Nonmetastatic Colorectal Cancer. JAMA Oncol 2019;5:1118-23. [Crossref] [PubMed]
- Dasari A, Morris VK, Allegra CJ, et al. ctDNA applications and integration in colorectal cancer: an NCI Colon and Rectal-Anal Task Forces whitepaper. Nat Rev Clin Oncol 2020;17:757-70. [Crossref] [PubMed]
- Moreira L, Balaguer F, Lindor N, et al. Identification of Lynch syndrome among patients with colorectal cancer. JAMA 2012;308:1555-65. [Crossref] [PubMed]
- Evrard C, Tachon G, Randrian V, et al. Microsatellite Instability: Diagnosis, Heterogeneity, Discordance, and Clinical Impact in Colorectal Cancer. Cancers (Basel) 2019;11:1567. [Crossref] [PubMed]
- Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology 2010;138:2073-2087.e3. [Crossref] [PubMed]
- Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
- André T, Shiu KK, Kim TW, et al. Pembrolizumab in Microsatellite-Instability-High Advanced Colorectal Cancer. N Engl J Med 2020;383:2207-18. [Crossref] [PubMed]
- Petrelli F, Ghidini M, Cabiddu M, et al. Microsatellite Instability and Survival in Stage II Colorectal Cancer: A Systematic Review and Meta-analysis. Anticancer Res 2019;39:6431-41. [Crossref] [PubMed]
- Wang B, Li F, Zhou X, et al. Is microsatellite instability-high really a favorable prognostic factor for advanced colorectal cancer? A meta-analysis. World J Surg Oncol 2019;17:169. [Crossref] [PubMed]
- Sobocińska J, Kolenda T, Teresiak A, et al. Diagnostics of Mutations in MMR/EPCAM Genes and Their Role in the Treatment and Care of Patients with Lynch Syndrome. Diagnostics (Basel) 2020;10:786. [Crossref] [PubMed]
- Bonadona V, Bonaïti B, Olschwang S, et al. Cancer risks associated with germline mutations in MLH1, MSH2, and MSH6 genes in Lynch syndrome. JAMA 2011;305:2304-10. [Crossref] [PubMed]
- Doerner J. Risk of Metachronous Colorectal Cancer in Lynch Syndrome: Who Needs an Extended Resection? Surgeries 2022;3:185-91. [Crossref]
- Anele CC, Adegbola SO, Askari A, et al. Risk of metachronous colorectal cancer following colectomy in Lynch syndrome: a systematic review and meta-analysis. Colorectal Dis 2017;19:528-36. [Crossref] [PubMed]
- Lam AK, Chan SS, Leung M. Synchronous colorectal cancer: clinical, pathological and molecular implications. World J Gastroenterol 2014;20:6815-20. [Crossref] [PubMed]
- Sepulveda AR, Hamilton SR, Allegra CJ, et al. Molecular Biomarkers for the Evaluation of Colorectal Cancer: Guideline From the American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and the American Society of Clinical Oncology. J Clin Oncol 2017;35:1453-86. [Crossref] [PubMed]
- Nakano K, Yamamoto H, Fujiwara M, et al. Clinicopathologic and Molecular Characteristics of Synchronous Colorectal Carcinoma With Mismatch Repair Deficiency. Am J Surg Pathol 2018;42:172-82. [Crossref] [PubMed]
- Vyas M, Firat C, Hechtman JF, et al. Discordant DNA mismatch repair protein status between synchronous or metachronous gastrointestinal carcinomas: frequency, patterns, and molecular etiologies. Fam Cancer 2021;20:201-13. [Crossref] [PubMed]
- Cereda M, Gambardella G, Benedetti L, et al. Patients with genetically heterogeneous synchronous colorectal cancer carry rare damaging germline mutations in immune-related genes. Nat Commun 2016;7:12072. [Crossref] [PubMed]
- Hu H, Chang DT, Nikiforova MN, et al. Clinicopathologic features of synchronous colorectal carcinoma: A distinct subset arising from multiple sessile serrated adenomas and associated with high levels of microsatellite instability and favorable prognosis. Am J Surg Pathol 2013;37:1660-70. [Crossref] [PubMed]
- Bae JM, Cho NY, Kim TY, et al. Clinicopathologic and molecular characteristics of synchronous colorectal cancers: heterogeneity of clinical outcome depending on microsatellite instability status of individual tumors. Dis Colon Rectum 2012;55:181-90. [Crossref] [PubMed]
- Ryoo SB, Heo S, Lim Y, et al. Personalised circulating tumour DNA assay with large-scale mutation coverage for sensitive minimal residual disease detection in colorectal cancer. Br J Cancer 2023;129:374-81. [Crossref] [PubMed]
- Chen K, Shields MD, Chauhan PS, et al. Commercial ctDNA Assays for Minimal Residual Disease Detection of Solid Tumors. Mol Diagn Ther 2021;25:757-74. [Crossref] [PubMed]
- Malla M, Loree JM, Kasi PM, et al. Using Circulating Tumor DNA in Colorectal Cancer: Current and Evolving Practices. J Clin Oncol 2022;40:2846-57. [Crossref] [PubMed]
- Jesinghaus M, Pfarr N, Kloor M, et al. Genetic heterogeneity in synchronous colorectal cancers impacts genotyping approaches and therapeutic strategies. Genes Chromosomes Cancer 2016;55:268-77. [Crossref] [PubMed]
- Morris VK, Yothers G, Kopetz S, et al. NRG-GI005 (COBRA): phase II/III study of circulating tumor DNA as a predictive biomarker in adjuvant chemotherapy in patients with stage II colon cancer. J Clin Oncol 2020;38:TPS261. [Crossref]
- Taïeb J, Benhaim L, Laurent Puig P, et al. Decision for adjuvant treatment in stage II colon cancer based on circulating tumor DNA: The CIRCULATE-PRODIGE 70 trial. Dig Liver Dis 2020;52:730-3. [Crossref] [PubMed]
- Tie J, Cohen J, Lahouel K, et al. Adjuvant chemotherapy guided by circulating tumor DNA analysis in stage II colon cancer: The randomized DYNAMIC trial. J Clin Oncol 2022;40:LBA100. [Crossref]
- Folprecht G, Reinacher-Schick A, Tannapfel A, et al. Circulating tumor DNA-based decision for adjuvant treatment in colon cancer stage II evaluation: (CIRCULATE-trial) AIO-KRK-0217. J Clin Oncol 2020;38:TPS273. [Crossref]


