
SPP1 promotes tumor progression in esophageal carcinoma by activating focal adhesion pathway
Highlight box
Key findings
• Secreted phosphoprotein 1 (SPP1) activates focal adhesion kinase (FAK)/Erk pathway in esophageal carcinoma (ESCA), thus promoting tumor progression.
What is known and what is new?
• SPP1 is considered as a hub gene in ESCA and is negatively associated with disease-free survival in ESCA.
• SPP1 promotes tumor progression in ESCA by activating FAK/Erk pathway.
What is the implication, and what should change now?
• FAK is a potential therapeutic target to overcome tumor recurrence and metastasis of ESCA.
• Optimization of the current strategies in the treatment of ESCA.
Introduction
Esophageal carcinoma (ESCA) is characterized by a complex pathogenesis and ranked as the tenth most common cancer and the sixth leading cause of cancer-related deaths worldwide (1). Despite the progress achieved in the diagnosis and therapeutic regimens, the 5-year survival rate remains poor in patients with ESCA (2-4). Recurrence and metastasis are the major obstacles that affect the therapeutic efficacy and clinical outcomes for patients with ESCA (5). Hence, there is an urgent need to unveil the molecular mechanisms involved in the recurrence and metastasis of ESCA, which will help to screen and identify the potential therapeutic targets and pave new ways to enhance the clinical therapeutic efficacy of ESCA treatment.
Secreted phosphoprotein 1 (SPP1; known as osteopontin) is a kind of secreted acidic protein (6), which is commonly expressed in osteocytes, macrophages, and endothelial cells, etc. (7-10) and regulates various biological functions, including bone remodeling, chemotaxis, cell activation, and apoptosis (6,11-15). Under pathological conditions, SPP1 is highly expressed in several types of cancers such as colorectal cancer, gastric cancer, liver cancer, pancreatic cancer and breast cancer, to name a few (16,17). In these tumors, SPP1 was reported to be associated with tumor proliferation, invasion, and stem-like behavior (17-19). In addition, SPP1 has been considered as a diagnostic and prognostic marker for gastric cancer, liver cancer, and breast cancer (17,20-23).
The reports on the roles of SPP1 in ESCA are very limited. Thus far, only two published reports revealed the roles of SPP1 in ESCA. One was published in 2015, which revealed that exogenous SPP1 treatment promoted the migration of ESCA cells (24). The other was published in 2022, which revealed that SPP1 was involved in radiation resistance through JAK2/STAT3 pathway in ESCA (25). Based on integrated bioinformatics analysis, SPP1 is considered as a hub gene (26-28), which is highly expressed in ESCA samples and is negatively associated with disease-free survival (DFS) in ESCA (28-31). Thus, the exact roles and the underlying mechanisms of SPP1 in ESCA require further exploration.
In the preliminary study, we found that genes in ESCA tissues with high expression of SPP1 were enriched in the focal adhesion pathway based on gene set enrichment analysis (GSEA). In the current study, we aim to examine the roles of SPP1 on focal adhesion, and elucidate the potential mechanisms, in order to optimize the therapeutic strategies and improve the clinical prognosis for patients with ESCA. We present this article in accordance with the MDAR reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-302/rc).
Methods
Bioinformatic analysis
The Cancer Genome Atlas (TCGA; http://www.cancergenome.nih.gov/) was used for GSEA to screen the enriched pathways in ESCA with a high expression of SPP1. The Kaplan-Meier plotter (http://www.kmplot.com/) was used to analyze the effect of SPP1 on clinical survival. UALCAN (https://ualcan.path.uab.edu/index.html) was used to analyze the correlations between SPP1 expression and clinicopathological characteristics.
Cell culture
Human ESCA cell lines (Eca-109 Cat#: ZQ0234, KYSE-410 Cat#: ZQ0928, and TE-1 Cat#: ZQ0235) were obtained from Zhong Qiao Xin Zhou Biotech (Shanghai, China). All cell lines were cultured in RPMI-1640 medium (Cat#: 350-030; Wisent, Nanjing, China) supplemented with 10% fetal bovine serum (Cat#: 087-150; Wisent) and 1% penicillin/streptomycin (Cat#: PC0315; ProteinBio, Nanjing, China).
Stable cell line construction
Lentivirus coding SPP1 short hairpin RNA (shRNA) was constructed by GeneChem (Shanghai, China) (5'-ACGAGTCAGCTGGATGACC-3'; control sequence: 5'-ATTGCGTTCGCAGTAATCT-3'). Cells (2×104/well) were seeded in 24-well plates and were infected with lentiviruses [multiplicity of infection (MOI) =50]. Stable cell clones were selected for 4–7 days using 0.5 µg/mL puromycin (Cat#: HY-K1057; MCE, NJ, USA).
Quantitative polymerase chain reaction (qPCR)
Total RNA was extracted from cells with TRIzol. Total RNA was reverse transcribed using a reverse transcription kit (Cat#: PE5402; ProteinBio) according to the manufacturer’s instructions. The SYBR Green qPCR Supermix (Cat#: PS2024; ProteinBio) was used for qPCR and the 2−ΔΔCt method was used to calculate the RNA levels.
The sequences of the primers for qPCR are as follows [SPP1: forward 5'-CTCCATTGACTCGAACGACTC-3', reverse 5'-CAGGTCTGCGAAACTTCTTAGAT-3'; glyceraldehyde-3-phosphate dehydrogenase (GAPDH): forward 5'-TGTGGGCATCAATGGATTTGG-3', reverse 5'-ACACCATGTATTCCGGGTCAAT-3'].
Cell counting kit-8 (CCK8)
Cells (1×103/well) were seeded in 96-well plates and incubated with complete medium at 37 ℃ with 5% CO2 for 24, 48, 72, and 96 h. Ten µL of CCK8 reagent (Cat#: HY-K0301; MCE) and 90 µL of complete medium were then mixed and added to each well. After 2 h of incubation at 37 ℃, the optical density (OD) values at 450 nm were detected.
Western blotting
The protein extracts were separated on 10% gels via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to 0.45 nm polyvinylidene difluoride membranes. After blocking, the membranes were incubated with specific primary antibodies as follows: GAPDH (Cat#: GTX627408; Genetex, San Antonio, TX, USA; 1:5,000), SPP1 (Cat#: 30200-1-AP; Proteintech, Wuhan, China; 1:1,000), focal adhesion kinase (FAK; Cat#: A11131; Abclonal, Wuhan, China; 1:1,000), p-FAK (Cat#: AP1447; Abclonal; 1:1,000), Erk1/2 (Cat#: A4782; Abclonal; 1:1,000), p-Erk1/2 (Cat#: RK05775; Abclonal; 1:1,000). After washing, the membranes were incubated with secondary antibodies (Cat#: AS080; HRP-conjugated anti-mouse/anti-rabbit antibody; Abclonal; 1:20,000). Finally, the proteins were visualized with super enhanced chemiluminescence (ECL) substrate kit (Cat#: PECLS09; ProteinBio).
Clone formation assay
A total of 500–1,000 cells were seeded in six-well plates, and the medium was replaced every 3–4 days. After 10–14 days, the colonies were fixed with 4% paraformaldehyde (PFA) for 10 min, and stained with 0.1% crystal violet solution for 20 min.
Wound-healing assay
The cells were seeded in six-well plates and cultured to confluence. The 200 µL pipette tip was used to produce the wound. The cells were cultured in serum-free medium after removing the detached cells with PBS. The wound images were collected at 0 and 12 h under a microscope.
Transwell assay
The cell invasion assay was performed as follows: the upper chambers were coated with Matrigel (Cat#: 356234; BD Biosciences, Franklin, NJ, USA), then cells (20,000–40,000 cells/well) in serum-free medium were seeded into the upper chamber (Cat#: 3422; Corning Inc., Corning, NY, USA), and complete medium was added to the lower chamber. After a 24-h culture, the cells in the top membrane were removed with cotton swabs. The cells on the lower membrane surface were fixed with 4% PFA and stained with 0.1% crystal violet.
Adhesion assay
Cells (1×104) were seeded in 96-well plates which were precoated with 10 µg/mL collagen (MCE) for 2 h at 37 ℃. After incubating at 37 ℃ for 1 h, cells that did not adhere to the plates were washed away with PBS. Adherent cells were fixed with 4% PFA, then stained with 0.1% crystal violet.
Cell spreading assay and immunofluorescence
Cells (2×104) were seeded in 2-cm dishes which were precoated with 10 µg/mL collagen (Cat#: HY-P70018; MCE) for 2 h at 37 ℃ followed by blocking with 1% BSA for 1 h. After culturing for 1 h, the cells were fixed, permeabilized and blocked sequentially. Then, the cells were incubated with the primary antibody of FAK (Cat#: A11131; Abclonal; 1:500) overnight at 4 ℃ followed by Cy3 conjugated anti-rabbit secondary antibody (Cat#: AS007; Abclonal; 1:300) for 1 h at room temperature. Finally, the cells were counterstained using YF488-Phalloidin (Cat#: PY0488; ProteinBio) and 4',6-diamidino-2-phenylindole (DAPI).
Statistical analysis
Statistical analysis was performed using GraphPad Prism (version 9.0). Quantitative data are expressed as mean ± standard deviation (SD) and analyzed using the Student’s t-test. A P<0.05 is considered statistically significant.
Results
SPP1 is upregulated in ESCA tissues and positively associated with disease progression and poor prognosis in patients with ESCA
We analyzed the gene expression profile in ESCA based on TCGA database. A total of 4,049 differentially expressed genes (2,636 up-regulated differentially expressed genes and 1,413 down-regulated differentially expressed genes) were identified under the criteria of fold change (FC) ≥2 and false discovery rate (FDR) <0.01 (Figure 1A). Among these differentially expressed genes, SPP1 was upregulated in ESCA tissues compared to the adjacent tissues (log2FC =4,473, P<0.001) (Figure 1B).
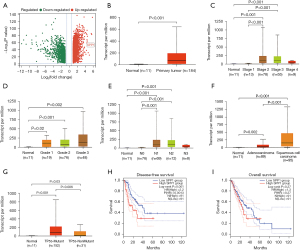
In the subcategory analysis, we found that SPP1 expression was relatively higher in stage 2 or 3 ESCA, which was higher than that in stage 1 (P<0.001) and normal tissues (P<0.001) (Figure 1C), while no differences were found among tumor grade in ESCA (Figure 1D). There was a higher expression of SPP1 in ESCA with N1 and N2 lymph node metastasis than that without lymph node metastasis (both P<0.001) (Figure 1E). In addition, the expression of SPP1 in squamous cell carcinoma subtype of ESCA was higher than that in adenocarcinoma subtype (P<0.001) (Figure 1F). The expression of SPP1 in ESCA with TP53 mutation was higher than that without TP53 mutation (P=0.006) (Figure 1G).
Survival analysis showed that there was a negative correlation between SPP1 expression and DFS (log-rank P=0.001) (Figure 1H), but no correlation was found between SPP1 expression and overall survival (OS) (log-rank P=0.27) (Figure 1I) in patients with ESCA, indicating that high expression of SPP1 predicts a poor clinical prognosis in patients with ESCA.
SPP1 promoted the malignant behaviors of ESCA cells
We then examined the expression of SPP1 in three ESCA cell lines and found that SPP1 was highly expressed in ESCA cells (Figure 2A). TE-1 and KYSE-410 were chosen for further study due to the relatively higher expression of SPP1. Result from the immunofluorescence assay confirmed that SPP1 was localized mainly in the cytoplasm of ESCA cells (Figure 2B).
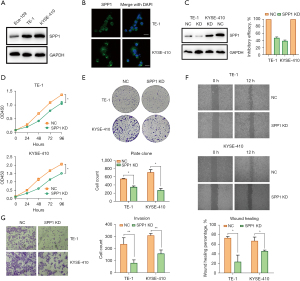
To examine the role of SPP1 on the malignance of ESCA, loss-of-function assay was performed by infecting TE-1 and KYSE-410 cells with lentivirus coding SPP1 shRNA to establish SPP1-knockdown cell lines. Western blotting and qPCR confirmed that SPP1 was significantly knocked down in these two cells (Figure 2C). CCK8 (Figure 2D) and plate clone (Figure 2E) assays showed that the knockdown of SPP1 repressed the cell proliferative capacities compared to the control cells. In addition, wound healing assay revealed that the knockdown of SPP1 reduced the migratory capacities of ESCA cells (Figure 2F), and transwell assay indicated that the knockdown of SPP1 inhibited the invasive capacities of ESCA cells (Figure 2G).
SPP1 promoted the malignant behaviors by activating focal adhesion of ESCA cells
To understand the mechanisms of SPP1 involved in the malignant progression of ESCA, GSEA was conducted with dataset from TCGA, and the results showed that the genes in ESCA tissues with a high expression of SPP1 were enriched in several pathways (Figure 3A). Among them, the focal adhesion pathway [normalized enrichment score (NES) =1.772, P=0.02] (Figure 3B) piqued our interest due to its close relation with tumor malignant progression. To verify this, we conducted cell adhesion assay and found that the knockdown of SPP1 in ESCA cells inhibited the ability of cells to adhere to collagen (Figure 3C). We further examined the morphology and location of focal adhesion by staining cells with focal adhesion marker (FAK) and observed that obvious pseudopods on the leading edge of the plasma membrane in control cells were almost obscure when SPP1 was knocked down in ESCA cells (Figure 3D). In addition, we found that the knockdown of SPP1 dramatically repressed the expression of p-FAK and p-Erk in ESCA cells (Figure 3E).
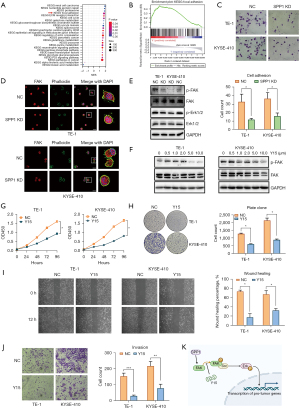
Y15 is a specific inhibitor of FAK. To further verify whether SPP1 regulates tumor progression in ESCA by activating focal adhesion, we treated ESCA cells with Y15. The results showed that Y15 inhibited FAK autophosphorylation in TE-1 at a dose equal or over 5.0 µM and in KYSE-410 at a dose equal or over 1.0 µM (Figure 3F) and dramatically inhibited proliferative (Figure 3G,3H), migratory (Figure 3I), and invasive (Figure 3J) capacities of ESCA cells.
Discussion
In the current study, we explored the roles of SPP1 in ESCA as well as the underlying mechanisms. To our knowledge, this is the first study that confirms that SPP1 is closely related to the focal adhesion pathway and regulates tumor progression by activating FAK/Erk pathway in ESCA. Furthermore, our findings reveal that FAK inhibitors are potential therapeutics in the management of ESCA.
In this study, SPP1 expression was positively correlated with clinical stage and lymph node metastasis and TP53 mutation. In addition, the expression of SPP1 in squamous cell carcinoma subtype of ESCA was higher than that in adenocarcinoma subtype. These findings indicate that SPP1 be used as a potential biomarker for patient risk stratification and local regional metastasis in ESCA.
We then confirmed that the knockdown of SPP1 inhibited the malignant behaviors of ESCA cells including cell proliferation, migration, and invasion, which was consistent with a previous report (31). Then, focal adhesion was identified as a vital pathway involved in the malignance of ESCA. Our study was the first evidence to prove that SPP1 exerts an oncogenic role in ESCA by activating the focal adhesion pathway. It is well-known that focal adhesion contains several proteins such as FAK and Src (32,33), and is closely associated with tumor growth and metastasis. FAK is a highly auto-phosphorylated protein and participates in cell proliferation, adhesion, and migration (34) by regulating cytoskeleton dynamics and cell movement. In the current study, we verified that SPP1can activate focal adhesion in ESCA cells, evidenced by decreased levels of p-FAK, obscure pseudopodiums, and less FAK on the leading edge of the periphery in SPP1-knockdown cells.
It has been reported that phosphorylated FAK is involved in several signaling pathways. Phosphorylated FAK in association with Src can further regulate PI3K/Akt, p53, and Erk signaling pathway (35-37). In the current study, we verified that SPP1 promoted cell proliferation, migration, and invasion by activating FAK/Erk signaling.
Based on the above results, FAK plays a vital role in tumor progression of ESCA. Thus, FAK-targeted strategies are promising to overcome the recurrence and metastasis of ESCA. To date, several FAK-targeted approaches have been developed, such as the knockdown of FAK using siRNA, antisense oligonucleotides, etc. Although these approaches dramatically inhibit tumor growth (38-40), toxicity in vivo is a big obstacle that hinders the clinical application of these approaches. Small-molecule agents possess good spatial dispersion, druggability, and pharmacokinetic properties, which are great advantages in drug development. Thus, the development of FAK-targeted small molecule agents seems promising. Y15 is a kind of FAK inhibitor, which can inhibit FAK autophosphorylation. In our study, we found that Y15 repressed cell proliferation, migration, and invasion in ESCA cells, indicating that targeting FAK is a promising strategy to overcome the malignance of ESCA.
It has been reported that the combination of FAK inhibitors with other therapeutics is more effective than either alone (41). For example, Y15 combined with gemcitabine was more effective in suppressing pancreatic cancer cell growth in a mouse model than either alone (42). In clinical trials, the efficacy data are limited. In a phase I clinical trial, the safety of FAK inhibitor (PF-04554878) in combination with pembrolizumab or gemcitabineto treat pancreatic cancer was evaluated, and although the drug tolerability is satisfactory, no partial or complete response was observed (43). A phase II clinical trial (NCT01870609) evaluated PF-04554878 in the treatment of pleural mesothelioma after first-line chemotherapy (44), but no benefit on DFS, OS or quality of life was achieved. To date, several combined strategies are under clinical evaluation for treatment of advanced solid cancers, and with more efficacy data published, the true potential of single or combinatorial therapies continues to be explored.
There are some limitations in this study. Firstly, the findings of this study need to be further confirmed in animal models, especially the anti-tumoral role of Y15 in ESCA. Secondly, clinical samples should be collected in our institution to verify the results from public databases. Lastly, in-depth mechanisms need to be explored to identify potential targets and develop targeted agents, which may be combined with Y15 as novel therapeutics in the treatment of ESCA.
Conclusions
The current study proposes a novel regulatory mechanism of SPP1 in the malignance of ESCA. SPP1 promotes tumor cell proliferation, invasion, and migration in ESCA, partly by initiating the formation of focal adhesion through FAK/Erk pathway (Figure 3K). The findings achieved in this study indicated that FAK and downstream molecules are potential molecular targets for ESCA therapy.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-302/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-302/dss
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-302/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-302/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Rustgi AK, El-Serag HB. Esophageal carcinoma. N Engl J Med 2014;371:2499-509. [Crossref] [PubMed]
- Shevde LA, Samant RS. Role of osteopontin in the pathophysiology of cancer. Matrix Biol 2014;37:131-41. [Crossref] [PubMed]
- Rooney MS, Shukla SA, Wu CJ, et al. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015;160:48-61. [Crossref] [PubMed]
- Ying J, Huang HH, Zhang MM, et al. Up-regulation of SOCS4 promotes cell proliferation and migration in esophageal squamous cell carcinoma. Transl Cancer Res 2021;10:2416-27. [Crossref] [PubMed]
- Koroknai V, Szász I, Jámbor K, et al. Expression pattern of osteopontin isoforms in malignant melanoma cell lines. Clin Transl Sci 2024;17:e13694. [Crossref] [PubMed]
- Kadoglou NPE, Khattab E, Velidakis N, et al. The Role of Osteopontin in Atherosclerosis and Its Clinical Manifestations (Atherosclerotic Cardiovascular Diseases)-A Narrative Review. Biomedicines 2023;11:3178. [Crossref] [PubMed]
- Kubota A, Frangogiannis NG. Macrophages in myocardial infarction. Am J Physiol Cell Physiol 2022;323:C1304-24. [Crossref] [PubMed]
- Denhardt DT, Noda M, O'Regan AW, et al. Osteopontin as a means to cope with environmental insults: regulation of inflammation, tissue remodeling, and cell survival. J Clin Invest 2001;107:1055-61. [Crossref] [PubMed]
- Fortunato F, Ferlini A. Biomarkers in Duchenne Muscular Dystrophy: Current Status and Future Directions. J Neuromuscul Dis 2023;10:987-1002. [Crossref] [PubMed]
- Choi ST, Kim JH, Kang EJ, et al. Osteopontin might be involved in bone remodelling rather than in inflammation in ankylosing spondylitis. Rheumatology (Oxford) 2008;47:1775-9. [Crossref] [PubMed]
- Qin YC, Yan X, Yuan XL, et al. Osteopontin promotes gastric cancer progression phosphatidylinositol-3-kinase/protein kinase B/mammalian target of rapamycin signaling pathway. World J Gastrointest Oncol 2023;15:1544-55. [Crossref] [PubMed]
- Burdo TH, Wood MR, Fox HS. Osteopontin prevents monocyte recirculation and apoptosis. J Leukoc Biol 2007;81:1504-11. [Crossref] [PubMed]
- Digifico E, Erreni M, Mannarino L, et al. Important functional role of the protein osteopontin in the progression of malignant pleural mesothelioma. Front Immunol 2023;14:1116430. [Crossref] [PubMed]
- Le JM, Wu JH, Jaw FS, et al. The effect of bone remodeling with photobiomodulation in dentistry: a review study. Lasers Med Sci 2023;38:265. [Crossref] [PubMed]
- Moorman HR, Poschel D, Klement JD, et al. Osteopontin: A Key Regulator of Tumor Progression and Immunomodulation. Cancers (Basel) 2020;12:3379. [Crossref] [PubMed]
- Zhao H, Chen Q, Alam A, et al. The role of osteopontin in the progression of solid organ tumour. Cell Death Dis 2018;9:356. [Crossref] [PubMed]
- Ho NT, Lin SW, Lee YR, et al. Osteopontin Splicing Isoforms Contribute to Endometriotic Proliferation, Migration, and Epithelial-Mesenchymal Transition in Endometrial Epithelial Cells. Int J Mol Sci 2022;23:15328. [Crossref] [PubMed]
- Bandopadhyay M, Bulbule A, Butti R, et al. Osteopontin as a therapeutic target for cancer. Expert Opin Ther Targets 2014;18:883-95. [Crossref] [PubMed]
- Nedjadi T, Ahmed ME, Ansari HR, et al. Identification of SPP1 as a Prognostic Biomarker and Immune Cells Modulator in Urothelial Bladder Cancer: A Bioinformatics Analysis. Cancers (Basel) 2023;15:5704. [Crossref] [PubMed]
- Briones-Orta MA, Avendaño-Vázquez SE, Aparicio-Bautista DI, et al. Osteopontin splice variants and polymorphisms in cancer progression and prognosis. Biochim Biophys Acta Rev Cancer 2017;1868:93-108.A.
- Cao DX, Li ZJ, Jiang XO, et al. Osteopontin as potential biomarker and therapeutic target in gastric and liver cancers. World J Gastroenterol 2012;18:3923-30. [Crossref] [PubMed]
- Walaszek K, Lower EE, Ziolkowski P, et al. Breast cancer risk in premalignant lesions: osteopontin splice variants indicate prognosis. Br J Cancer 2018;119:1259-66. [Crossref] [PubMed]
- Lin J, Myers AL, Wang Z, et al. Osteopontin (OPN/SPP1) isoforms collectively enhance tumor cell invasion and dissemination in esophageal adenocarcinoma. Oncotarget 2015;6:22239-57. [Crossref] [PubMed]
- Wang M, Sun X, Xin H, et al. SPP1 promotes radiation resistance through JAK2/STAT3 pathway in esophageal carcinoma. Cancer Med 2022;11:4526-43. [Crossref] [PubMed]
- Song Y, Wang X, Wang F, et al. Identification of four genes and biological characteristics of esophageal squamous cell carcinoma by integrated bioinformatics analysis. Cancer Cell Int 2021;21:123. [Crossref] [PubMed]
- Feng Z, Qu J, Liu X, et al. Integrated bioinformatics analysis of differentially expressed genes and immune cell infiltration characteristics in Esophageal Squamous cell carcinoma. Sci Rep 2021;11:16696. [Crossref] [PubMed]
- Tang L, Chen Y, Peng X, et al. Identification and Validation of Potential Pathogenic Genes and Prognostic Markers in ESCC by Integrated Bioinformatics Analysis. Front Genet 2020;11:521004. [Crossref] [PubMed]
- Zhang H, Zhong J, Tu Y, et al. Integrated Bioinformatics Analysis Identifies Hub Genes Associated with the Pathogenesis and Prognosis of Esophageal Squamous Cell Carcinoma. Biomed Res Int 2019;2019:2615921. [Crossref] [PubMed]
- Wang Y, Lu Y, Xu W, et al. Prognostic value of osteopontin expression in esophageal squamous cell carcinoma: A meta-analysis. Pathol Res Pract 2019;215:152571. [Crossref] [PubMed]
- Li M, Wang K, Pang Y, et al. Secreted Phosphoprotein 1 (SPP1) and Fibronectin 1 (FN1) Are Associated with Progression and Prognosis of Esophageal Cancer as Identified by Integrated Expression Profiles Analysis. Med Sci Monit 2020;26:e920355. [PubMed]
- Zhang S, Lai T, Su X, et al. Linc00662 m(6)A promotes the progression and metastasis of pancreatic cancer by activating focal adhesion through the GTF2B-ITGA1-FAK pathway. Am J Cancer Res 2023;13:1718-43. [PubMed]
- Liu W, Bloom DA, Cance WG, et al. FAK and IGF-IR interact to provide survival signals in human pancreatic adenocarcinoma cells. Carcinogenesis 2008;29:1096-107. [Crossref] [PubMed]
- Fife CM, McCarroll JA, Kavallaris M. Movers and shakers: cell cytoskeleton in cancer metastasis. Br J Pharmacol 2014;171:5507-23. [Crossref] [PubMed]
- Li S, Wu H, Wang F, et al. Enhanced Bone Regeneration through Regulation of Mechanoresponsive FAK-ERK1/2 Signaling by ZINC40099027 during Distraction Osteogenesis. Int J Med Sci 2024;21:137-50. [Crossref] [PubMed]
- Patel A, Sabbineni H, Clarke A, et al. Novel roles of Src in cancer cell epithelial-to-mesenchymal transition, vascular permeability, microinvasion and metastasis. Life Sci 2016;157:52-61. [Crossref] [PubMed]
- Chen T, Wang J, Li M, et al. Genistein Inhibits Proliferation and Metastasis in Human Cervical Cancer Cells through the Focal Adhesion Kinase Signaling Pathway: A Network Pharmacology-Based In Vitro Study in HeLa Cells. Molecules 2023;28:1919. [Crossref] [PubMed]
- Tan X, Yan Y, Song B, et al. Focal adhesion kinase: from biological functions to therapeutic strategies. Exp Hematol Oncol 2023;12:83. [Crossref] [PubMed]
- Zhang Z, Li J, Jiao S, et al. Functional and clinical characteristics of focal adhesion kinases in cancer progression. Front Cell Dev Biol 2022;10:1040311. [Crossref] [PubMed]
- Golubovskaya VM, Zheng M, Zhang L, et al. The direct effect of focal adhesion kinase (FAK), dominant-negative FAK, FAK-CD and FAK siRNA on gene expression and human MCF-7 breast cancer cell tumorigenesis. BMC Cancer 2009;9:280. [Crossref] [PubMed]
- Golubovskaya VM. Targeting FAK in human cancer: from finding to first clinical trials. Front Biosci (Landmark Ed) 2014;19:687-706. [Crossref] [PubMed]
- Golubovskaya VM, Nyberg C, Zheng M, et al. A small molecule inhibitor, 1,2,4,5-benzenetetraamine tetrahydrochloride, targeting the y397 site of focal adhesion kinase decreases tumor growth. J Med Chem 2008;51:7405-16. [Crossref] [PubMed]
- Wang-Gillam A, Lockhart AC, Tan BR, et al. Phase I study of defactinib combined with pembrolizumab and gemcitabine in patients with advanced cancer. J Clin Oncol 2018;36:2561. [Crossref]
- Fennell DA, Baas P, Taylor P, et al. Maintenance Defactinib Versus Placebo After First-Line Chemotherapy in Patients With Merlin-Stratified Pleural Mesothelioma: COMMAND-A Double-Blind, Randomized, Phase II Study. J Clin Oncol 2019;37:790-8. [Crossref] [PubMed]


