
The roles and mechanisms of APOL1 in the development of colorectal cancer
Highlight box
Key findings
• The apolipoprotein L1-runt-related transcription factor 1 (APOL1-RUNX1) signaling axis mediated the Wnt/β-catenin pathway, which regulated the biological behavior of colorectal cancer (CRC) cells.
What is known and what is new?
• APOL1 was highly expressed in CRC. Silencing APOL1 inhibited the ability of CRC cells to proliferate, migrate and invade, and suppressed the proliferation and growth of transplanted tumors in vivo.
• Overexpression of RUNX1 reversed the inhibitory effect of silencing APOL1 on epithelial-mesenchymal transition (EMT), proliferation, migration, invasion and Wnt/β-catenin pathway in CRC cells.
What is the implication, and what should change now?
• APOL1 is a key factor in EMT, proliferation, migration and invasion of CRC cells. This provided new clues and directions for the study of CRC-related molecular mechanisms.
Introduction
A prevalent malignant tumor in the digestive system, colorectal cancer (CRC) is one of the leading causes of cancer-related mortality worldwide (1). The prognosis is dismal for patients with advanced CRC, and the 5-year survival rate has not improved greatly despite the rapid development of surgical treatment, chemotherapy, radiation therapy, and targeted therapy (2). Therefore, investigating the molecular mechanisms behind the onset and progression of CRC and identifying novel regulatory targets for enhancing the therapy regimen for CRC patients are crucial to improving therapeutic outcomes.
Numerous studies have confirmed the link between apolipoproteins a range of biological processes, such as tumor growth, immunological response, and inflammatory response. Recent research has demonstrated that apolipoprotein is directly involved in the development, apoptosis, migration, and invasion of tumor cells and can be exploited as a biomarker for the early diagnosis and prognosis of a variety of malignancies (3). Apolipoprotein L1 (APOL1) is involved in lipid transport and metabolism as a secreted high-density lipoprotein (HDL). APOL1 is located on chromosome 22q12.3 and binds to apolipoprotein A1 (APOA1) (4). Numerous malignancies, such as bladder cancer, small-cell lung cancer, and hepatocellular carcinoma, have high expression levels of APOL1 (5-7). In contrast, renal cancer tissues and cells exhibit a considerable downregulation of APOL1 expression (8). It is still unknown, therefore, how APOL1 functions and how it causes CRC.
In this study, we thus thoroughly investigated the biological role of APOL1 in CRC and preliminarily explored the potential molecular mechanisms by which APOL1 exerts its biological function in CRC. We present this article in accordance with the ARRIVE reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-275/rc).
Methods
Clinical specimens
The study comprised 20 patients who were undergoing radical CRC surgery at the Affiliated Hospital of Guangdong Medical University. The postoperative cancer tissue was recovered, along with adjacent normal tissue that was 3 cm away from the cancer tissue. Quantitative real-time polymerase chain reaction (qRT-PCR) was used detect the gene expression of APOL1. None of the patients had received radiation therapy or chemotherapy before the operation. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of Affiliated Hospital of Guangdong Medical University (No. PJ2020-090) and informed consent was taken from all the patients.
Cell culture
Shanghai iCell Bioscience Inc. (Shanghai, China) provided the human normal colon epithelial cells NCM460 (iCell-h373) and CRC cell lines [HCT116 (iCell-h071) and SW1116 cells (iCell-h201)]. The culture of the cells was conducted in Dulbecco’s Modified Eagle Medium. When there was at least 80% confluence, the cell fusion was deemed complete. With cells in the logarithmic development phase, further experimental studies were conducted.
Cell transfection
Shanghai Scigrace Biotech Co., Ltd. (Shanghai, China) supplied the shRNA-APOL1 (sh-APOL1) and its nonspecific negative control (NC) oligos, plasmid cloning DNA (pcDNA)-runt-related transcription factor 1 (RUNX1) and pcDNA-NC. Cells were cultured and transfected with Lipofectamine 2000. sh-NC and sh-APOL1 were transfected into HCT116 and SW1116 cells. After transfection, the cells expressing sh-APOL1-1, sh-APOL1-2 and sh-APOL1-3 were tested with qRT-PCR (Table 1).
Table 1
| Gene | Sequence |
|---|---|
| shAPOL1-1 | CCAGAGAGCAGTATCTTTATT |
| shAPOL1-2 | GGACAACCTTGCAAGACAAAT |
| shAPOL1-3 | GTGAGCTTGAGGATAACATAA |
The sh-NC, sh-APOL1, pcDNA-NC, and pcDNA-RUNX1 were transfected into CRC cells lines. The cells were divided into four groups: the sh-NC, sh-APOL1, sh-APOL1 + pcDNA-NC, and sh-APOL1 + pcDNA-RUNX1 groups. This allowed us to investigate how the interaction between the APOL1 and RUNX1 proteins affects HCT116 and SW1116 cells.
SKL2001 (40 µM) is dissolved in dimethyl sulfoxide (DMSO) before use and needs to be added to the culture plate 4 h before transfection.
Cell Counting Kit 8 (CCK8) assay
The transfected cells were cultivated at a density of 2×104 cells per well, which is a common cell density used for cell proliferation. Each well was then seeded with 1 mL of culture medium to provide sufficient nutrition and space for cell growth. Each well received a 100-µL addition of cell culture media. The cell viability in the plate was assessed 48 hours after the cells were cultured at room temperature. Subsequently, 10 µL of CCK8 assay reagent was added to each well and treated at 37 ℃ for 2 hours. The optical density at 450 nm was measured by an enzyme marker.
Clonogenic assay
Transfected cells were injected into six-well plates at appropriate concentrations to allow cell growth and expansion within the wells. After 2 weeks of culture, 1×103 cells were plated in each well. During culture, the medium was switched every 3 days. The cultivation was stopped when cells became discernible. After one wash with phosphate-buffered saline (PBS), the cells were fixed with 1 mL of 4% paraformaldehyde per well. Each well received 1 mL of crystal violet dye solution, which was then stained for 10–20 min. After the PBS had dried, the cells were cleaned multiple times and photographed. After the number of cell clones in each well was calculated three times, the average value was determined.
Transwell assay
The cells were cultured in a medium for full growth and proliferation. The cells were transferred to a six-well plate with 2´105 cells per well. Following the addition of 300 µL of cell suspension to the upper chamber and 450 µL of RPMI-1640 medium to the lower chamber of a Transwell device that contained a membrane coated with matrix gel, the device was incubated for 48 hours in a CO2 incubator. At the end of the culture, cells were fixed at the bottom of the chamber membrane with methanol for 20 minutes for cell visualization and counting. After staining with 0.1% crystal violet was completed for 20 minutes, a PBS wash was applied for 20 minutes. Under an inverted microscope (Leica, Wetzlar, Germany), five randomly selected fields were counted.
Wound healing assay
We inoculated 2´105 cells into six-well plates and let them sit in the medium for 24 hours. Subsequently, we examined the cells under a microscope and recorded the initial breadth of the scratches. The scratch breadth was measured and recorded one more time after a 24-hour incubation period.
Animals and groups
Fourteen Female BALB/c-nu mice, 4–5 weeks of age and weighing between 18–22 g, were used in the experiments. These specific-pathogen-free-grade mice were provided by the Tumor Control Center, Sun Yat-sen University (Guangzhou, China). The mice at the facility received food and clean drinking water. Animal experiments were conducted at West China Hospital of Sichuan University. Animal experiments were performed under a project license (No. 20230215021) granted by ethics board of West China Hospital of Sichuan University, in compliance with national guidelines for the care and use of animals. A protocol was prepared before the study without registration. Two groups, designated sh-APOL1 and sh-NC, were created from the mice, with each group consisting of 7 mice. Randomly assign mice into groups using the Rand function in Excel to generate randomization. The sh-NC or sh-APOL1 was transfected into SW1116 cells. Subcutaneous injection of 1×107 cells/mL of transfected SW1116 cells was then performed to create the tumorigenic nude mice model. Every 7 days for 3 weeks in a succession, the tumor volume was measured, and the mice were killed after 28 days. One mouse died prematurely, leading to the inability to collect data and the other mouse, which failed to develop cell tumorigenesis, was excluded from the study. Conduct subsequent testing on each group of 6 mice (n=6). This study, for the first time, conducted in vivo experimental assessment of the role of APOL1 in CRC, hence using a relatively small sample size. Therefore, the primary objective of the study was to collect basic experimental data to provide support for more complex experimental designs in the future.
Immunohistochemistry
As stated above, the tumor tissues were sectioned, paraffin-fixed, and then dehydrated using different alcohol percentages (75%, 85%, 95%, and 100%, volume/volume). Subsequently, the pieces were then blocked for 20 minutes with 3% H2O2 and washed three times in PBS for 5 minutes each. The slices were again washed in PBS and then blocked for 15 minutes using goat serum. The sections were then left overnight at 4 ℃ to be treated with a primary antibody against Ki-67 (ab15580; Abcam, Cambridge, UK). Goat anti-mouse immunoglobin G (IgG) heavy chain and light chain (ab150113; Abcam) was added to the sections for 2 hours at 37 ℃, and then the sections were stained for 5–7 minutes with 3,3’-diaminobenzidine (DAB). Following a 5-minute PBS washing, the slices underwent a second staining for 2 minutes with hematoxylin (Beyotime Biotechnology, Shanghai, China), a 1-minute treatment with fractionated alcohol in hydrochloric acid, and a 10-minute rinse under running water. After the slices were observed under an optical microscope (Leica) to determine the positive protein expression, they were naturally dried, sealed, and then photographed.
qRT-PCR analysis
Tumor tissues and CRC cells were treated with TRIzol reagent to extract RNA, which was followed by reverse transcription into complement DNA (cDNA). The cDNA served as the template for the qRT-PCR protocol. Denaturation took place for 5 minutes at 95 ℃, with 40 cycles of 95 ℃ for 15 seconds, 60 ℃ for 30 seconds, 72 ℃ for 40 seconds, and 55 ℃ for 15 seconds. Each primer sequence is listed in full in Table 2 (MBL Beijing Biotech Co., Ltd., Beijing, China). With the 2–ΔΔCt method, the messenger RNA (mRNA) level of APOL1 was determined after the experiment’s cycle threshold (Ct) values were estimated.
Table 2
| Gene | Primer | Primer sequence |
|---|---|---|
| APOL1 | APOL1-forward primer | 5'-TGGACTACGGAAAGAAGTGGT-3' |
| APOL1-reverse primer | 5'-CCTCCTTCAATTTGTCAAGGCTT-3' | |
| GAPDH | GAPDH-forward primer | 5'-AGTACCCCATTGAACACGGC-3' |
| GAPDH-reverse primer | 5'-TGCTTCACCACCTTCTTG-3' |
Coimmunoprecipitation
HCT116 and SW1116 cells were collected, rinsed with PBS, mixed with an appropriate amount of radioimmunoprecipitation assay (RIPA) buffer (including protease inhibitors), and cleaved on ice for 30 minutes. Centrifugation was used to extract the supernatant at 12,000 rpm for 20 minutes at 4 ℃. We removed a minute portion of supernatant for western blotting. After being mixed with 1 μg of the appropriate antibody, the leftover supernatant was incubated at 4 ℃ overnight. After 10 µL of protein A beads were removed, they were washed three times with the appropriate amount of RIPA buffer and centrifuged for 3 minutes at 3,000 rpm. The antibody and protein A beads were conjugated by adding 10 μL of pretreated protein A beads to the supernatant that was left overnight to be incubated at 4 ℃ for 24 hours. This mixture was then centrifuged at 3,000 rpm for 3 minutes at 4 ℃. After washing the beads three times in cold RIPA buffer, the supernatants were discarded. Then, 15 μL of 2× sodium dodecyl sulfate loading buffer was added, and the mixture was left to boil for 5 minutes. Finally, the binding protein was identified via western blotting.
Western blotting
To extract total protein from tumor tissues and CRC cells, RIPA buffer (Solarbio, Beijing, China) was used. A bicinchoninic acid (BCA) protein quantification kit (Solarbio) was used to calculate the protein content in each group. Following this, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to separate the proteins before they were transferred to polyvinylidene fluoride (PVDF) membranes. After 2 hours of blocking of the membrane with 5% skim milk, the membrane was incubated for overnight at 4 ℃ with anti-APOL1 (ab252218; Abcam), anti-RUNX1 (ab229482; Abcam), anti-β-catenin (ab32572; Abcam), anti-cyclin D1 (ab16663; Abcam), anti-E-cadherin (ab40772; Abcam), anti-vimentin (ab92547; Abcam), and anti-N-cadherin (ab76011; Abcam). Subsequently, the membrane was incubated at room temperature for 1 hour with the horseradish peroxidase (HRP) anti-rabbit IgG antibody (ab288151; Abcam). To observe protein bands, an improved chemiluminescence kit (Thermo Fisher Scientific, Waltham, MA, USA) was employed. The target protein bands were evaluated for gray-scale values using ImageJ software (US National Institutes of Health, Bethesda, MD, USA). The internal reference used was β-actin.
Statistical analysis
All experimental data were collected by a researcher unaware of the groupings. GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, CA, USA) was used to statistically analyze the data. The experimental results are expressed as the mean ± standard deviation. The two groups were compared using the t-test. Statistical significance was set at P<0.05.
Results
Expression of APOL1 in CRC tissue
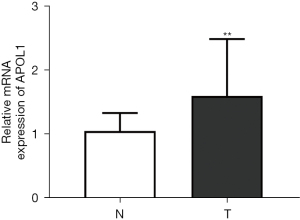
Clinical samples were used to examine APOL1 expression in CRC. The findings (Figure 1) demonstrated that APOL1 was highly expressed at the mRNA level in CRC tissues (1.60±0.89) compared to adjacent normal tissues (1.05±0.28, P=0.009).
Effect of APOL1 on the biological behavior of CRC cells
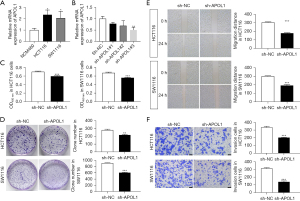
Compared to NCM460 cells (1.01±0.16), HCT116 cells (2.35±0.79, P=0.02) and SW1116 cells (2.06±0.72, P=0.04) had substantially greater APOL1 mRNA levels (Figure 2A). Only APOL1#3 mRNA expression (0.50±0.26, P=0.006) was less than 50% compared with the sh-NC group (1.00±0.10) according to the qRT-PCR data, suggesting that sh-APOL1#3’s interference efficiency was higher than 50% (Figure 2B). CCK8 showed (Figure 2C) that sh-APOL1 decreased the proliferative capacity of HCT116 (0.60±0.02, P<0.001) and SW1116 cells (0.56±0.01, P<0.001). This was also demonstrated by colony formation assay (Figure 2D), where sh-APOL1 decreased the proliferation ability of HCT116 (217.67±9.29, P=0.002) and SW1116 cells (607.33±13.80, P<0.001). The results of the wound healing (Figure 2E) indicated that sh-APOL1 reduced the migration ability of HCT116 (177.33±5.80, P<0.001) and SW1116 cells (190.5±11.72, P<0.001). The results of Transwell assays (Figure 2F) indicated that sh-APOL1 reduced the ability of HCT116 (203.33±4.73, P<0.001) and SW1116 cells (139.33±11.68, P<0.001) to invade.
Effect of APOL1 on tumor growth and metastasis in CRC tumor-bearing mice
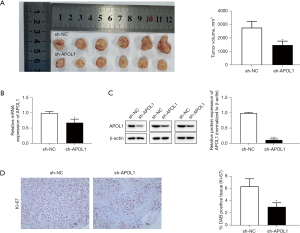
We investigated the effect of APOL1 on tumor formation and metastasis in CRC tumor-bearing mice. The sh-APOL1 group’s tumor volume (1,483.42±791.24, P=0.02) steadily dropped after 28 days compared to that of the sh-NC group (2,776.93±471.27) (Figure 3A). According to the results of qRT-PCR (Figure 3B) and Western blotting (Figure 3C), the mRNA level of APOL1 in the sh-APOL1 group (0.69±0.19, P=0.01) was lower than that in the sh-NC group (1.00±0.06), and the protein expression of APOL1 declined in the sh-APOL1 group (0.11±0.48, P<0.001) was smaller than that in the sh-NC group (6.35±1.26). The nuclear protein Ki-67 promoted the formation and development of malignant tumors. According to immunohistochemical examination, the sh-APOL1 group (2.99±0.69, P=0.02) had lower levels of positive Ki-67 expression (Figure 3D). Therefore, in mice with CRC tumors, APOL1 silencing inhibited the growth and spread of tumors.
Validation of the protein interaction of APOL1 with RUNX1
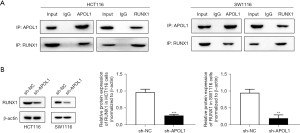
In recent years, bioinformatics analysis has been considered a functional tool for studying tumor mechanisms (9). The Harmonizome 3.0 website (https://maayanlab.cloud/Harmonizome/gene/APOL1) predicted that the APOL1-interacting protein was RUNX1. The coimmunoprecipitation results indicated that the APOL1 protein interacted with RUNX1 protein (Figure 4A). The results of the Western blotting demonstrated that RUNX1 protein expression in the HCT116 cells (0.28±0.38, P<0.001) and SW1116 cells (0.20±0.42, P<0.001) was greatly decreased by APOL1 silencing (Figure 4B).
RUNX1 overexpression affected the epithelial-mesenchymal transition (EMT) and biological behavior of CRC cells by regulating the Wnt/β-catenin pathway
One of the most extensively conserved signaling pathways is the Wnt-β-catenin pathway, which exerts a considerable influence in both tumor and embryonic development. It can contribute to tumor progression by triggering proliferative signals downstream (10,11). In HCT116 cells, RUNX1 (0.24±0.03, P<0.001), β-catenin (0.54±0.16, P=0.001), cyclin D1 (0.37±0.24, P=0.004), N-cadherin (0.33±0.03, P<0.001), and vimentin (0.31±0.10, P<0.001) down-regulated and E-cadherin (3.64±0.58, P<0.001) up-regulated in the sh-APOL1 group compared to the sh-APOL1 group (Figure 5A). In SW1116 cells, RUNX1 (0.16±0.07, P<0.001), β-catenin (0.16±0.09, P=0.001), cyclin D1 (0.35±0.20, P=0.003), N-cadherin (0.32±0.15, P=0.001), and vimentin (0.38±0.03, P<0.001) down-regulated and E-cadherin (5.61±1.13, P<0.001) up-regulated in the sh-APOL1 group compared to the sh-APOL1 group (Figure 5A). This suggests that silencing APOL1 inhibited Wnt-β-catenin pathway activation and the EMT process. In HCT116 (0.57±0.02; 151.50±13.94; 86.67±16.74, P<0.001) and SW1116 cells (0.55±0.02; 139.17±17.34; 106.67±9.61, P<0.001), sh-APOL1 reduced and proliferation, migration and invasion (Figure 5B-5D). The inhibitory effects of sh-APOL1 on EMT, proliferation, migration, and invasion of CRC cells were countered by the overexpression of RUNX1 (Figure 5A-5D). In HCT116 cells, RUNX1 (0.75±0.11, P<0.001), β-catenin (0.84±0.12, P=0.04), cyclin D1 (0.87±0.31, P=0.045), N-cadherin (0.70±0.09, P=0.01) and vimentin (0.62±0.14, P=0.045) up-regulated and E-cadherin (1.68±0.17, P<0.001) down-regulated in the sh-APOL1 + pcDNA-RUNX1 group compared to the sh-APOL1 + pcDNA-NC group (Figure 5A). In SW1116 cells, RUNX1 (0.38±0.08, P=0.001), β-catenin (0.81±0.27, P=0.02), cyclin D1 (0.64±0.27, P=0.03), N-cadherin (0.65±0.15, P=0.04) and vimentin (0.75±0.08, P<0.001) up-regulated and E-cadherin (2.58±0.55, P=0.002) down-regulated in the sh-APOL1 + pcDNA-RUNX1 group compared to the sh-APOL1 + pcDNA-NC group (Figure 5A). In HCT116 (0.62±0.01, P=0.002; 249.17±24.50, P<0.001; 131.00±6.25, P<0.001) and SW1116 cells (0.60±0.02, P=0.01; 216.00±23.82, P=0.01; 153.33±10.97, P=0.003), the ability of proliferation, migration and invasion was increased in the sh-APOL1 + pcDNA-RUNX1 group (Figure 5B-5D).
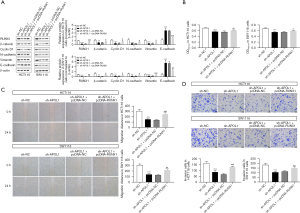
Silencing the APOL1-mediated Wnt-β-catenin pathway affected EMT, proliferation, migration, and invasion in CRC cells
In HCT116 (2.54±1.00, P=0.03; 0.48±0.19, P<0.001; 0.99±0.27, P=0.04; 0.99±0.17, P<0.001; 0.89±0.19, P=0.006; 0.71±0.09, P<0.001) and SW1116 cells (3.64±1.56, P=0.004; 0.56±0.13, P<0.001; 0.91±0.19, P=0.006; 0.89±0.09, P=0.001; 0.95±0.20, P=0.005; 0.56±0.27, P<0.001), there was a substantial decrease in the expression of E-cadherin and a significant rise in the expression of RUNX1, β-catenin, cyclin D1, N-cadherin and vimentin in the sh-APOL1 + SKL2001 group (Figure 6A). Biological behavior of HCT116 and SW1116 cells was higher in the sh-APOL1 + SKL2001 group compared to the sh-APOL1 group (Figure 6B-6D). In HCT116 (0.62±0.01, P=0.001; 228.00±12.01, P<0.001; 108.67±14.98, P=0.02) and SW1116 cells (0.61±0.01, P<0.001; 169.00±5.29, P=0.009; 146.33±18.90, P=0.02), the ability of proliferation, migration and invasion was increased in the sh-APOL1 + SKL2001 group (Figure 6B-6D). This suggests that APOL1 suppression facilitated the Wnt-β-catenin pathway, which regulated EMT and the biological behavior of CRC cells.
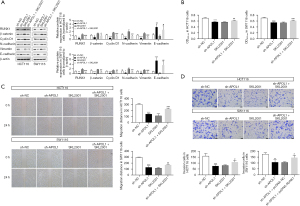
Discussion
A number of genes are altered as a result of common internal and external variables, both hereditary and environmental, that contribute to the development and spread of CRC (12,13). It has been discovered that the TP53, APC, and COX2 genes are intimately linked to CRC (14). However, since none of these genes can be used as a specific early diagnostic molecular indicator, they also cannot be used as therapeutic targets with precise efficacy. Thus, a thorough investigation of the molecular biological mechanisms underlying the development of CRC is essential for improving both the clinical therapy and molecular diagnosis of the disease, as well as for extending patient prognosis and quality of life (15,16).
Pancreatic cancer cells can proliferate when APOL1 is employed as an oncogene (17). Our study revealed that APOL1 was highly expressed in clinical tumor specimens. sh-APOL1 was transfected in HCT116 and SW1116 cells to determine the effect of APOL1 on the biological behavior of CRC cells. The findings showed that HCT116 and SW1116 cells’ capacity for invasion, migration, and proliferation were all suppressed by silencing APOL1. sh-APOL1 prevented tumor growth in the tumor-bearing mouse model. The most reliable molecular marker for assessing the proliferative activity of cancer cells in CRC is Ki-67, and sh-APOL1 in this study significantly reduced Ki-67 expression. These findings provide evidence for APOL1’s oncogenic function in CRC.
CRC is a disease of multiple genetic abnormalities, and gene expression is regulated by signaling pathways. Numerous investigations have revealed that the Wnt-β-catenin pathway is intimately associated with the progression of malignant tumors (18,19). The cyclin D1 is a direct participant in tumor cell cycle, proliferation regulation, growth, and deterioration (20). When the organism is abnormal, Wnt-β-catenin pathway induces β-catenin into the nucleus and promotes the transcription of downstream effector gene cyclin D1, ultimately affecting cell proliferation and migration (21). In order to prevent CRC invasion and metastasis, Cinobufacini suppresses EMT and the Wnt-β-catenin signaling pathway (20). Syntrophin beta 1 (SNTB1) controls the biological behavior of CRC cells through the Wnt-β-catenin pathway (22). The findings of our study demonstrated that sh-APOL1 downregulates β-catenin and cyclin D1, which in turn activates this pathway. Furthermore, the involvement of the Wnt/β-catenin pathway in mediating the EMT process and its significance in gastric, breast, lung, and CRC has also been established in earlier research (23-26). The expressions of N-cadherin and vimentin were both downregulated in this study, but E-cadherin expression was upregulated by APOL1 silencing, with the implication being that APOL1 silencing prevented the EMT process of CRC. Subsequently, the Wnt-β-catenin pathway activator (SKL2001) was used to further confirm the significance of this pathway in regulating EMT and to clarify the biological mechanisms by which APOL1 influences CRC cells. The inhibitory effect of APOL1 silencing on CRC cells’ EMT, proliferation, invasion, and migration was reversed after SKL2001 was used to silence APOL1. The influence of APOL1 on EMT and the biological behavior of CRC cells is likely mediated by the Wnt/β-catenin pathway.
The indirect and direct biological roles of RUNX1 include controlling angiogenesis, cancer metastasis, proliferation, and chemical resistance to anticancer medications (27). The function of RUNX1 has been extensively studied in multiple cancer types. Through attenuating RUNX1-induced transcriptional stimulation of the Warburg effect, YT521-B homology (YTH) domain-containing protein (YTHDC1)-mediated augmentation of miR-30d is able to suppress pancreatic carcinogenesis (28). By means of CRC’s stimulation of the Wnt-β-catenin pathway and EMT, RUNX1 has been demonstrated to enhance tumor spread (29). Bioinformatics analysis reveals a possible protein interaction between APOL1 and RUNX1. In this study, coimmunoprecipitation indicated that APOL1 could bind RUNX1 protein in HCT116 and SW1116 cells and that sh-APOL1 could downregulate RUNX1 protein expression. This suggests that APOL1 is able to bind to RUNX1 proteins and regulate their protein levels. Silencing APOL1 significantly inhibited the expression of RUNX1 and significantly impaired the EMT process, proliferative ability, migration and invasive behavior of CRC cells through the inhibition of the Wnt/β-catenin pathway.
Conclusions
In CRC cells, APOL1 played a crucial role in the EMT, proliferation, migration, and invasion. Furthermore, it was shown that the biological behavior of CRC cells was regulated by the APOL1-RUNX1 signaling axis, which mediated the Wnt-β-catenin pathway. These findings offer novel insights and directions in the study of the molecular mechanisms related to CRC. However, a limitation of this study is that the clinical sample may not adequately represent the diversity of all CRC patients, especially given the heterogeneity of CRC.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-275/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-275/dss
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-275/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-275/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of Affiliated Hospital of Guangdong Medical University (No. PJ2020-090) and informed consent was taken from all the patients. Animal experiments were performed under a project license (No. 20230215021) granted by ethics board of West China Hospital of Sichuan University, in compliance with national guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- Li J, Ma X, Chakravarti D, et al. Genetic and biological hallmarks of colorectal cancer. Genes Dev 2021;35:787-820. [Crossref] [PubMed]
- Ren L, Yi J, Li W, et al. Apolipoproteins and cancer. Cancer Med 2019;8:7032-43. [Crossref] [PubMed]
- Zhong F, Lu HP, Chen G, et al. The clinical significance of apolipoprotein L1 in head and neck squamous cell carcinoma. Oncol Lett 2020;20:377. [Crossref] [PubMed]
- Shi J, Yang H, Duan X, et al. Apolipoproteins as Differentiating and Predictive Markers for Assessing Clinical Outcomes in Patients with Small Cell Lung Cancer. Yonsei Med J 2016;57:549-56. [Crossref] [PubMed]
- Bharali D, Banerjee BD, Bharadwaj M, et al. Expression analysis of apolipoproteins AI & AIV in hepatocellular carcinoma: A protein-based hepatocellular carcinoma-associated study. Indian J Med Res 2018;147:361-8. [Crossref] [PubMed]
- Ma XL, Gao XH, Gong ZJ, et al. Apolipoprotein A1: a novel serum biomarker for predicting the prognosis of hepatocellular carcinoma after curative resection. Oncotarget 2016;7:70654-68. [Crossref] [PubMed]
- Hu CA, Klopfer EI, Ray PE. Human apolipoprotein L1 (ApoL1) in cancer and chronic kidney disease. FEBS Lett 2012;586:947-55. [Crossref] [PubMed]
- Chen Y, Qi C, Xia L, et al. Identification of novel genetic etiology and key molecular pathways for seminoma via network-based studies. Int J Oncol 2017;51:1280-90. [Crossref] [PubMed]
- Zhang Y, Wang X. Targeting the Wnt/β-catenin signaling pathway in cancer. J Hematol Oncol 2020;13:165. [Crossref] [PubMed]
- Yu F, Yu C, Li F, et al. Wnt/β-catenin signaling in cancers and targeted therapies. Signal Transduct Target Ther 2021;6:307. [Crossref] [PubMed]
- Xu D, Han S, Yue X, et al. METTL14 Suppresses Tumor Stemness and Metastasis of Colon Cancer Cells by Modulating m6A-Modified SCD1. Mol Biotechnol 2023; Epub ahead of print. [Crossref] [PubMed]
- Abdalla Abdelaziz MA, Nelson VK, Kumarasamy M, et al. Anticancer effect of polyphenolic acid enriched fractions from Grewia bracteata Roth on tumor cells and their p53 gene independent ROS mediated apoptosis in colon cancer cells. Toxicon 2023;233:107243. [Crossref] [PubMed]
- Shieh Y, Eklund M, Sawaya GF, et al. Population-based screening for cancer: hope and hype. Nat Rev Clin Oncol 2016;13:550-65. [Crossref] [PubMed]
- Sallinger K, Gruber M, Müller CT, et al. Spatial tumour gene signature discriminates neoplastic from non-neoplastic compartments in colon cancer: unravelling predictive biomarkers for relapse. J Transl Med 2023;21:528. [Crossref] [PubMed]
- Zheng Y, Wu J, Chen H, et al. KLF4 targets RAB26 and decreases 5-FU resistance through inhibiting autophagy in colon cancer. Cancer Biol Ther 2023;24:2226353. [Crossref] [PubMed]
- Lin J, Xu Z, Xie J, et al. Oncogene APOL1 promotes proliferation and inhibits apoptosis via activating NOTCH1 signaling pathway in pancreatic cancer. Cell Death Dis 2021;12:760. [Crossref] [PubMed]
- Yan R, Zhu H, Huang P, et al. Liquidambaric acid inhibits Wnt/β-catenin signaling and colon cancer via targeting TNF receptor-associated factor 2. Cell Rep 2022;38:110319. [Crossref] [PubMed]
- Liu J, Xiao Q, Xiao J, et al. Wnt/β-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal Transduct Target Ther 2022;7:3. [Crossref] [PubMed]
- Wang J, Cai H, Liu Q, et al. Cinobufacini Inhibits Colon Cancer Invasion and Metastasis via Suppressing Wnt/β-Catenin Signaling Pathway and EMT. Am J Chin Med 2020;48:703-18. [Crossref] [PubMed]
- Zheng CC, Liao L, Liu YP, et al. Blockade of Nuclear β-Catenin Signaling via Direct Targeting of RanBP3 with NU2058 Induces Cell Senescence to Suppress Colorectal Tumorigenesis. Adv Sci (Weinh) 2022;9:e2202528. [Crossref] [PubMed]
- Zhang H, Li Z, Jiang J, et al. SNTB1 regulates colorectal cancer cell proliferation and metastasis through YAP1 and the WNT/β-catenin pathway. Cell Cycle 2023;22:1865-83. [Crossref] [PubMed]
- Tang Q, Chen J, Di Z, et al. TM4SF1 promotes EMT and cancer stemness via the Wnt/β-catenin/SOX2 pathway in colorectal cancer. J Exp Clin Cancer Res 2020;39:232. [Crossref] [PubMed]
- Tian S, Peng P, Li J, et al. SERPINH1 regulates EMT and gastric cancer metastasis via the Wnt/β-catenin signaling pathway. Aging (Albany NY) 2020;12:3574-93. [Crossref] [PubMed]
- Zhu L, Tian Q, Gao H, et al. PROX1 promotes breast cancer invasion and metastasis through WNT/β-catenin pathway via interacting with hnRNPK. Int J Biol Sci 2022;18:2032-46. [Crossref] [PubMed]
- Pan J, Fang S, Tian H, et al. lncRNA JPX/miR-33a-5p/Twist1 axis regulates tumorigenesis and metastasis of lung cancer by activating Wnt/β-catenin signaling. Mol Cancer 2020;19:9. [Crossref] [PubMed]
- Lin TC. RUNX1 and cancer. Biochim Biophys Acta Rev Cancer 2022;1877:188715. [Crossref] [PubMed]
- Hou Y, Zhang Q, Pang W, et al. YTHDC1-mediated augmentation of miR-30d in repressing pancreatic tumorigenesis via attenuation of RUNX1-induced transcriptional activation of Warburg effect. Cell Death Differ 2021;28:3105-24. [Crossref] [PubMed]
- Li Q, Lai Q, He C, et al. RUNX1 promotes tumour metastasis by activating the Wnt/β-catenin signalling pathway and EMT in colorectal cancer. J Exp Clin Cancer Res 2019;38:334. [Crossref] [PubMed]


