
Evaluating the mitochondrial structure and gene expression profile of regenerated liver tissues in mice after 85% partial hepatectomy
Highlight box
Key findings
• There were significant changes about mitochondrial morphology and related genes in regenerated tissues after 85% partial hepatectomy (PH) in mice.
What is known and what is new?
• Proliferating cell nuclear antigen (PCNA) positive rate was significantly increased after 85% PH in mice.
• Mitochondrial swelling, a reduction in the number of cristae, cristae disintegrated were showed in 85% PH group, and 30 differentially expressed genes (DEGs) were found by the reverse transcription-polymerase chain reaction (RT-PCR) chip, 29 DEGs were upregulated genes.
What is the implication, and what should change now?
• Mitochondria are the key organelles of energy supply. Mitochondrial respiratory chain proteins are highly expressed during liver regeneration after 85% PH.
Introduction
Partial hepatectomy (PH) is widely acknowledged to be the optimal treatment method for liver cancer worldwide (1-3). PH facilitates an early diagnosis of liver cancer, thereby considerably increasing the rate of survival. According to the Chinese Society of Clinical Oncology (CSCO) Guidelines for the Diagnosis and Treatment of Primary Liver Cancer, hepatectomy is recommended for patients with stage Ia–IIa disease or certain patients at stages II and III. However, extensive hepatectomy can cause complications, leading to liver failure and death in severe cases (4). Our initial study suggested that the preservation of mitochondrial function in the remaining liver following PH is paramount for successful liver regeneration and survival and even has the potential to suppress liver failure (5). In addition, a study conducted in 2012 illustrated that mitochondrial dysfunction in liver transplantation adversely affects the survival rate of mice that have undergone PH (6). The direct observation of live mice revealed that >75% of mitochondria exhibited depolarization changes following small-for-size liver transplantation, decreasing adenosine triphosphate (ATP) supply by >70% and consequently hindering regeneration (7,8). The literature suggests that inadequate liver preservation can cause liver failure in clinical scenarios (9), the minimization of which can potentially enhance fat metabolism and alleviate liver failure (10). Research regarding the influence of changes in the respiratory chain complex protein on liver regeneration in mice is rather scarce. In addition, the role of energy metabolism in liver regeneration warrants further exploration.
After PH, the process of regeneration starts rapidly; at this time, mitosis and the ensuing inflammatory response require large amounts of energy, and inadequate energy supply can lead to liver failure (11). Mitochondria produce >90% of the energy required for hepatic metabolism; however, damage to mitochondrial DNA can severely affect liver regeneration (12,13). The abnormal expression of mitochondrial complex-associated proteins can lead to abnormal energy metabolism (14). The subunits of the mitochondrial respiratory chain complex are closely linked to endosomal structures. Thorough bioinformatics analyses can be used to elucidate the relationships among numerous differentially expressed genes (DEGs) and comprehensively and systematically analyze the effects of mitochondrial DEGs (mt-DEGs) on liver regeneration. In this study, we analyzed the differential expression of the genes of mitochondrial respiratory chain complex subunits in liver regeneration tissues after 85% PH hepatectomy and characterized the association between liver regeneration, mitochondrial morphology, and the participating genes. We present this article in accordance with the ARRIVE reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-243/rc).
Methods
Animal model
For this study, twenty 8- to 10-week-old male C57BL/6J mice (weighing 22–25 g) were purchased from the Specific Pathogen Free Center, Dalian Medical University, Dalian, China. The experiments were performed under a project license (approval No. AEE19042) granted by the Ethical Review of Animal Experimentation, Dalian Medical University, in compliance with institutional guidelines for the care and use of animals. The mice were housed at a temperature of 22–25 ℃ and a 12-hour light-dark cycle. Food and water were freely available. The mice were randomly divided into false-negative and 85% PH groups (n=10/group). Of the total number of mice, five were observed to determine the 7-day survival rate and 15 were collected for sample collection. The mice were administered anesthesia via the continuous inhalation of sevoflurane (Hengrui Pharmaceuticals, Chengdu, China). A median longitudinal incision 2–2.5 cm in length was made on the abdomen of the mice in the sham group to expose the entire epigastric region. The cut was left for 25 min, following which the abdomen was sutured. In the 85% PH model mice, the falciform ligament, porta hepatis, left lateral lobe, and right diaphragmatic ligament were cut using miniature scissors; the left lateral lobe, middle lobe, right lower lobe, and caudate lobe were resected sequentially, and only the right upper lobe was retained. Postoperative observation revealed no blood leakage from the cut surface of the liver and a normal blood supply to the residual liver. The abdominal cavity was sutured and the incision sterilized. Following surgery, the mice were incubated at 25 ℃ for 30–60 minutes, following which they were removed from the incubator for individual rearing. The mice were humanely killed via decapitation 24 hours after the surgery, following which tissues were collected and preserved or processed accordingly. Furthermore, considering the guideline for animal experiments of reduction, refinement, replacement, and response, we selected three mice in the 85% PH group at 24 hours for subsequent experiments.
Hematoxylin and eosin staining assay
Liver tissues were collected from sham and 85% PH mice after 24 hours, washed with saline, fixed in 4% paraformaldehyde, and embedded in paraffin for 48 hours. The liver tissues were them sliced into 5-µm-thick sections, dewaxed, and hydrated using a concentration gradient of anhydrous ethanol. The tissue sections were stained using hematoxylin and eosin (HE) according to the manufacturer’s instructions (Beyotime, Shanghai, China). Morphological examination was performed using a BX3-CBH microscope (Olympus, Tokyo, Japan) at 100× magnification.
Immunohistochemistry (IHC) assay
IHC assay was performed to determine the expression changes in proliferating cell nuclear antigen (PCNA) (Proteintech, Wuhan, China) in the tissue arrays of the sham and 85% PH groups. The tissue sections were treated with varying degrees of alcohol and then incubated in citric acid and goat serum for antigenic repair and blocking according to the manufacturer’s protocol (ZSGB-BIO, Beijing, China). The tissue sections were then incubated in 1:2,000 PCNA primary antibody at 4 ℃ overnight. Subsequently, the tissue sections were incubated with the secondary antibody at room temperature for 1 h. The images were visualized using a DAB kit (ZSGB-BIO, Beijing, China) under a BX3-CBH microscope (Olympus, Tokyo, Japan). ImageJ software was used to determine the PCNA-positive cells at 200× magnification.
Transmission electron microscope (TEM) assay
TEM was used to observe mitochondrial morphology and microstructure. The fresh liver tissue was sectioned into 1×1×1 mm3 pieces, fixed with 2.5% glutaraldehyde (Solarbio, Beijing, China), and then fixed with 1% osmic acid at room temperature in the dark. The liver tissue was rinsed with phosphate-buffered saline, dehydrated with alcohol and then acetone, embedded using an embedding agent, polymerized at 60 ℃ for 48 hours, sliced into 60- to 80-nm ultrathin sections using an ultrathin microtome, stained with 150-mesh copper mesh and 2% uranium acetate-saturated alcohol solution in the dark, washed with 70% alcohol and pure water, stained with 2.6% lead citrate solution, and then dried at room temperature. Mitochondria were observed using TEM (JEM-1200Flash, JEOL, Beijing, China), and images were collected in a 100-nm field of view.
Screening for DEGs
After the operation, liver tissues were collected from three mice, and mitochondrial gene detection was performed based on the SABiosciences PCR chip platform (Aksomics, Shanghai, China). The DEGs were screened according to a P value of <0.05 and |fold change (FC)| of ≥1.5, with FC ≥1.5 indicating upregulation and FC ≤−1.5 indicating downregulation.
Volcano plot and correlation analyses
Volcano plot and correlation analyses were performed using “Volcano plot” and “Corrplot” tools in Hiplot Pro (https://hiplot.com.cn/) (15) based on the R v. 4.1.0 software packages “ggpubr”, “corrplot”, and “ggplot2” (The R Foundation for Statistical Computing).
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of DEGs
GO functional annotation, including biological process (BP), cellular component (CC), and molecular function (MF), and KEGG pathway enrichment analyses were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) database (https://david.ncifcrf.gov/summary.jsp) (16). The top 10 most enriched GO items and KEGG pathways were visualized via the R software packages “clusterProfiler” and “pheatmap”.
Protein-protein interaction (PPI) networks
The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (http://string-db.org/) was used to identify the PPIs (17). DEGs were used to construct the PPI network, with a high-confidence (0.7) in the mousenetwork text. Subsequently, Cytoscape 3.8.0 software was used to draw the PPI network map of the DEGs, and the cyto-Hubba plug-in was used to screen the core genes.
Quantitative real-time polymerase chain reaction assay (qRT-PCR)
qRT-PCR was performed using Applied Biosystems a Step One Plus Real-Time-PCR System (Thermo Fisher Scientific, Massachusetts, USA). Total RNA was extracted from each group of specimens or cells using TRIzol reagent (Invitrogen, Massachusetts, USA) according to the manufacturer’s instruction. Complement DNA (cDNA) reverse transcription was performed using EasyScript RT/RI Enzyme Mix and gDNA Eraser kit (TransGen Biotech, Beijing, China). The reagents used in the qRT-PCR assay were from the TransStart Tip Green qPCR SuperMix (ROX) (TransGen Biotech, Beijing, China) reagent kit. GAPDH was used as the internal reference for ATP5A1, ATP5B, ATP5O, CYC1, COX4I1, and UQCRC2. The primers are presented in Table 1. The relative intensities were quantified using the 2−△△CT method. Then, we set the expression level to 1 in the Sham group.
Table 1
| Symbol | Primers | |
|---|---|---|
| ATP5A1 | Forward primer | 5'CTGCATCTACGTCGCGATTG |
| Reverse primer | 5'CCGGAGTAGGGAGCCAAGTAC | |
| ATP5B | Forward primer | 5'AGTTGCTGAGGTCTTCACGG |
| Reverse primer | 5'CTTTGCCACGGCTTCTTC | |
| ATP5O | Forward primer | 5'TCATTTGGGTTTGACCTACAGCC |
| Reverse primer | 5'AGACCTGAACAGGGGGCCTTAC | |
| CYC1 | Forward primer | 5'CAGAAGTCTTGGAGTATGATG |
| Reverse primer | 5'ATCAACAACATCTTGAGACC | |
| COX4I1 | Forward primer | 5'CAGCGGTGGCAGAATGTTG |
| Reverse primer | 5'ACACCGAAGTAGAAATGGCTCTCT | |
| UQCRC2 | Forward primer | 5'ATTTCCCTGTAAGTGCCT |
| Reverse primer | 5'CTTGGGTTCTTGTTCTGA | |
| GAPDH | Forward primer | 5'GGTGAAGGTCGGTGTGAACG |
| Reverse primer | 5'CTCGCTCCTGGAAGATGGTG |
Statistical analyses
Statistical analyses were performed using R software packages “limma” and “clusterProfiler” for bioinformatics analyses. GraphPad Prism 7 software (GraphPad Software) was used to record the dates on which IHC and qRT-PCR were performed. We used t-test statistical method for the above two tools.and the P value of <0.05 was considered statistically significant.
Results
HE, IHC, and TEM morphological analyses
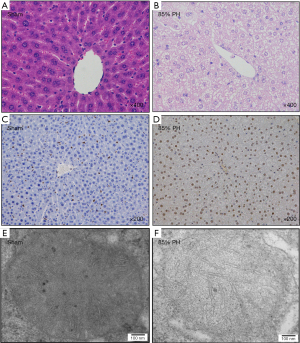
HE staining was performed to evaluate the regenerated liver tissues after 24 hours. In the sham group, the hepatic cords were distributed regularly in a radial pattern, morphology and the structure of the hepatic cells were normal, size of the hepatic cells was consistent, nuclear staining was light, nuclear cytoplasm was evenly distributed, and the boundary of the central vein and hepatic sinusoid were clear (Figure 1A). Conversely, in the regenerated liver tissues in the 85% PH group, the distribution of hepatic cords was irregular, the morphology and structure of the hepatic cells were irregular, the size of the hepatic cells was inconsistent, nuclear staining and cytoplasm were uneven, the boundary of the central vein and hepatic sinusoid was unclear, and the cells exhibited serious edema and pronounced steatosis with structural rupture, indicating structural abnormalities (Figure 1B). The PCNA positivity rate significantly increased 24 h after 85% PH in C57BL/6 mice compared with that in the sham group (Figure 1C,1D). TEM revealed mitochondrial swelling, and cristae disintegration in the 85% PH group (Figure 1E,1F).
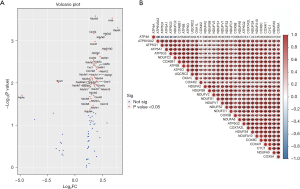
Mt-DEGs identified and visual analyses
The SABiosciences PCR chip identified 30 mt-DEGs (P<0.05 and |FC| ≥1.5). Among them, 29 genes were upregulated while ATP4A was downregulated. The DEGs were identified via SABiosciences PCR chips (Table 2). To show the DEGs more intuitively, a volcano plot was developed and the gene names were labeled (P<0.05; Figure 2). The correlation analyses of the mt-DEGs indicated significant positive correlations between ATP5A and ATP5O, NDUFA10 and COX6C, and COX4I1 and CYC1. COX4I2 and COX5B exhibited a weak correlation with the other genes.
Table 2
| Symbol | RefSeq | FC | P value | Description |
|---|---|---|---|---|
| ATP4A | NM_018731 | −4.81 | <0.05 | ATPase, H+/K+ exchanging, gastric, alpha polypeptide |
| ATP5A1 | NM_007505 | 2.28 | <0.01 | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit |
| ATP5B | NM_016774 | 2.24 | <0.01 | ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit |
| ATP5G1 | NM_007506 | 2.22 | <0.01 | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit c1 |
| ATP5G2 | NM_026468 | 2.01 | <0.01 | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C2 |
| ATP5G3 | NM_175015 | 2.78 | <0.001 | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 |
| ATP5O | NM_138597 | 1.90 | <0.01 | ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| ATP6V0A2 | NM_011596 | 2.84 | <0.01 | ATPase, H+ transporting, lysosomal V0 subunit A2 |
| COX4I1 | NM_009941 | 1.83 | <0.01 | Cytochrome c oxidase subunit IV isoform 1 |
| COX4I2 | NM_053091 | 1.70 | <0.05 | Cytochrome c oxidase subunit IV isoform 2 |
| COX5A | NM_007747 | 1.74 | <0.001 | Cytochrome c oxidase, subunit Va |
| COX5B | NM_009942 | 1.77 | <0.01 | Cytochrome c oxidase, subunit Vb |
| COX6B1 | NM_025628 | 1.88 | <0.01 | Cytochrome c oxidase, subunit VIb polypeptide 1 |
| COX6C | NM_053071 | 1.78 | <0.05 | Cytochrome c oxidase, subunit VIc |
| COX7A2L | NM_009187 | 1.97 | <0.01 | Cytochrome c oxidase subunit VIIa polypeptide 2-like |
| CYC1 | NM_025567 | 1.72 | <0.01 | Cytochrome c-1 |
| NDUFA10 | NM_024197 | 1.99 | <0.05 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 10 |
| NDUFA2 | NM_010885 | 1.53 | <0.05 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2 |
| NDUFA5 | NM_026614 | 1.88 | <0.001 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| NDUFA8 | NM_026703 | 2.25 | <0.001 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8 |
| NDUFB8 | NM_026061 | 1.51 | <0.05 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex 8 |
| NDUFC1 | NM_025523 | 1.67 | <0.05 | NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1 |
| NDUFC2 | NM_024220 | 2.19 | <0.01 | NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2 |
| NDUFS1 | NM_145518 | 1.64 | <0.05 | NADH dehydrogenase (ubiquinone) Fe–S protein 1 |
| NDUFS2 | NM_153064 | 1.73 | <0.01 | NADH dehydrogenase (ubiquinone) Fe–S protein 2 |
| NDUFS4 | NM_010887 | 1.95 | <0.01 | NADH dehydrogenase (ubiquinone) Fe–S protein 4 |
| NDUFV1 | NM_133666 | 1.76 | <0.01 | NADH dehydrogenase (ubiquinone) flavoprotein 1 |
| NDUFV2 | NM_028388 | 1.56 | <0.01 | NADH dehydrogenase (ubiquinone) flavoprotein 2 |
| OXA1L | NM_026936 | 2.03 | <0.01 | Oxidase assembly 1-like |
| UQCRC2 | NM_025899 | 2.26 | <0.05 | Ubiquinol Cytochrome c reductase core protein 2 |
DEGs, differentially expressed genes; RT-PCR, reverse transcription-polymerase chain reaction; FC, fold change.
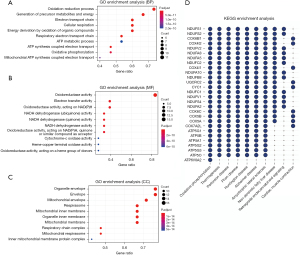
GO and KEGG enrichment analyses
GO and KEGG pathway enrichment analyses using the DAVID online analysis tool were performed to better understand the 29 upregulated genes, and the biological annotation of the upregulated genes with a P value of <0.05 was obtained. The top 10 most enriched GO terms are presented in Figure 3A-3C and included oxidation-reduction process, precursor metabolite and energy generation, electron transport chain (ETC), cellular respiration, energy derivation via the oxidation of organic compounds in the BP category; oxidoreductase activity, electron transfer activity, oxidoreductase activity acting on nicotinamide-adenine dinucleotide phosphate (NADPH), NADH dehydrogenase (ubiquinone) activity, and NADH dehydrogenase (quinone) activity in the MF category; and cadherin binding involved in organelle envelope, envelope, mitochondrial envelope, respirasome, and inner mitochondrial membrane in the CC category. The top 10 most enriched KEGG pathways are shown in Figure 3D, including oxidative phosphorylation, thermogenesis, Parkinson disease, prion disease, and nonalcoholic fatty liver disease (NAFLD).
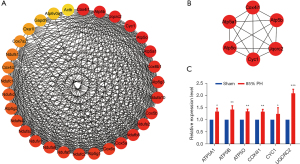
PPI analyses and qRT-PCR validation
All the DEGs were uploaded to the STRING database to construct PPI networks. Overall, 31 genes exhibited high confidence, and a complex network of DEGs was constructed using Cytoscape 3.8.0 (Figure 4A). The six most significant genes obtained using the cyto-Hubba plug-in were COX4I1, ATP5B, UQCRC2, CYC1, ATP5O, and ATP5A1 (Figure 4B). The expression levels of the genes were significantly different (P<0.05) as confirmed via qRT-PCR and consistent with those obtained via the SABiosciences PCR chip (Figure 4C).
Discussion
Postoperative liver failure is a severe complication with an incidence ranging from 0.7% to 9.1% (18). Predicting liver reserve has been an area of clinical focus. One study of an 85% PH model reported that all rats survived 72 h following model establishment (19). In other studies, mice demonstrated similarly exceptional survival rates after 85% PH, with a notable critical point for liver regeneration within 24–48 h after the surgery (20,21). In our study, no significant necrosis was observed 24 h after 85% PH, whereas PCNA exhibited a significantly significant positivity rate, suggesting a critical role of energy reserve in cell proliferation after PH (22).
Maintaining cellular functions and supporting cell proliferation require adequate energy supply. ATP is a key player in regenerative signaling and can regulate regeneration via hepatocyte purinoceptors when released after PH (23). Mitochondria are the predominant organelles in hepatocytes and primarily govern energy metabolism. After 85% PH, the most noticeable alteration in mitochondria is swelling, which is likely tied to the oxidative stress triggered by cholesterol accumulation in hepatocytes (24). The disintegration of mitochondrial cristae occurs because of Mic60 core protein deletion in the mitochondrial contact site and the cristae organization system (25), and these phenomena were consistent with observation in our previous study, in which Mic60 was underexpressed during 85% PH (26). Mic60 maintains mitochondrial integrity and function, and its deficiency can induce mitochondrial dysfunction (27).
The top ten GO (cellular component, CC) included inner mitochondrial membrane, inner organelle membrane, and respiratory chain complex, among others. The protein complexes embedded in the inner mitochondrial membrane, which include complex I (NADH dehydrogenase), complex II (succinate dehydrogenase), complex III (Cytochrome bc1 complex), complex IV (Cytochrome c oxidase), and ATP synthase, contribute to the ETC (28). Among the 29 DEGs that assemble into these complexes, the most substantial change was observed in complex I during 85% PH (Figure 5). NADH dehydrogenase (ubiquinone), NADH dehydrogenase (quinone), NADH dehydrogenase, and oxidoreductase, which act on NADPH, exhibited remarkable enrichment in the top 10 MF terms. Complex I comprises 45 subunits and serves as the main entry point for electrons into the ETC (29,30). Conversely, complex IV comprises 13 subunits and provides an outlet from the ETC (31,32). For ATP synthesis, the ATP synthase merges two components, F0 and F1, to stimulate proton reflux (33). Regarding the BP terms, ETC, oxidation–reduction, and ATP synthesis coupled to electron transport are critical for providing a large amount of ATP, which is necessary for liver regeneration (34,35). Twenty-eight DEGs partake in the oxidative phosphorylation reaction, which involves the respiratory chain complexes I, III, and IV and produces ATP. Abnormalities in mitochondrial thermogenesis may contribute to obesity and type 2 diabetes (36). Thermogenesis is also an energy expenditure mechanism and can potentially serve as an anti-obesity treatment (37,38). The KEGG pathway results of this study indicated that 28 DEGs participate in thermogenesis while simultaneously helping reduce steatosis and enhancing liver protection. However, thermogenesis might also consume the energy pivotal for cell proliferation. Hence, the role of thermogenesis in liver regeneration warrants further investigation. NAFLD is instigated by steatosis due to the presence of minimal residual liver following PH and can cause liver impairment, such as cirrhosis (39). The involvement of NAFLD suggests that lipid accumulation might potentially harm the regeneration of liver after 85% PH. In KEGG enrichment analyses, 21 DEGs were found to be associated with the genes involved in the NAFLD pathway, and their close link with mitochondria suggests that in the future, targeting at the mitochondrial level could mitigate liver damage induced by NAFLD and protect the regenerating liver (39).
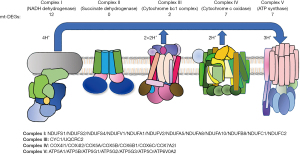
The six core genes identified via the cytoHubba plug-in primarily participate in cytochrome c oxidase and ATP syntheses, thereby significantly contributing to the energy supply for liver regeneration. ATP5A1, a complex V subunit alpha histone, has been implicated in neonatal mitochondrial encephalopathy (40). ATP5A1 overexpression inhibits tumor cell growth and NLRP3 inflammasome activation, maintaining mitochondrial function (41,42). The overexpression of ATP5B, a β polymorphism of the F1 subunit of complex V, can stimulate cell proliferation and contributes to tumor progression (43,44). Of note, ATP5B promotes the progression and metastasis of hepatocellular carcinoma (HCC) (45), which may prevent the treatment of HCC with PH. The overexpression of ATP5O leads to mitochondrial dysfunction, which can inhibit tumor growth and metastasis; however, the relationship between ATP5O and ATP production remains unclear (46). When downregulated, CYC1, a vital protein in the heme subunit of complex III, can enhance cell proliferation by lowering apoptosis levels, efficiently blocking metastasis in cancer cells (47,48). However, the upregulation of CYC1 may not significantly increase ATP production (49). COX4I1, functioning as a structural protein in the subunit 4 of complex IV, has a low expression in patients with insulin resistance and type 2 diabetes (50-52). UQCRC2, a core protein in complex III, is associated with spermatogenesis and male fertility (53) and can be used to treat liver failure (54). In addition, its low expression can obstruct cell proliferation, and its overexpression may encourage cancer development (55). Despite these findings, the precise mechanism remains unclear.
Conclusions
The 85% PH model promotes mitochondrial complex protein expression, with the alteration in complex I being the most significant, as it provides energy for liver regeneration. However, steatosis may inhibit liver regeneration. Key enrichments include the oxidation-reduction process, electron transfer activity, inner mitochondrial membrane processes, and others. In addition, COX4I1, ATP5B, UQCRC2, CYC1, ATP5O, and ATP5A1 were identified as potential core hub genes in the mitochondrial respiratory chain in this model. Overall, this study identified the changes of respiratory chain complex proteins in liver regeneration tissue after 85% PH, which may provide a basis for the study of the effect of energy metabolism on a hepatectomy model.
Acknowledgments
We would like to thank Shanghai Tengyun Biotechnology Co. for developing the Hiplot platform (https://hiplot.com.cn/).
Funding: This work was supported by grants from
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-243/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-243/dss
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-243/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-243/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The experiments were performed under a project license (approval No. AEE19042) granted by the Ethical Review of Animal Experimentation, Dalian Medical University, in compliance with institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Thayer D, Noda C, Charalel R, et al. Survival comparison of hepatocellular carcinoma patients treated with radioembolization versus nonoperative/interventional treatment. J Comp Eff Res 2018;7:343-56. [Crossref] [PubMed]
- Lee S, Kang TW, Cha DI, et al. Radiofrequency ablation vs. surgery for perivascular hepatocellular carcinoma: Propensity score analyses of long-term outcomes. J Hepatol 2018;69:70-8. [Crossref] [PubMed]
- Yagi S, Hirata M, Miyachi Y, et al. Liver Regeneration after Hepatectomy and Partial Liver Transplantation. Int J Mol Sci 2020;21:8414. [Crossref] [PubMed]
- Shang HT, Bao JH, Zhang XB, et al. Comparison of Clinical Efficacy and Complications Between Laparoscopic Partial and Open Partial Hepatectomy for Liver Carcinoma: A Meta-Analysis. J Laparoendosc Adv Surg Tech A 2019;29:225-32. [Crossref] [PubMed]
- Rehman H, Sun J, Shi Y, et al. NIM811 prevents mitochondrial dysfunction, attenuates liver injury, and stimulates liver regeneration after massive hepatectomy. Transplantation 2011;91:406-12. [Crossref] [PubMed]
- Shi Y, Rehman H, Ramshesh VK, et al. Sphingosine kinase-2 inhibition improves mitochondrial function and survival after hepatic ischemia-reperfusion. J Hepatol 2012;56:137-45. [Crossref] [PubMed]
- Liu Q, Rehman H, Krishnasamy Y, et al. Amphiregulin stimulates liver regeneration after small-for-size mouse liver transplantation. Am J Transplant 2012;12:2052-61. [Crossref] [PubMed]
- Liu Q, Rehman H, Krishnasamy Y, et al. Role of inducible nitric oxide synthase in mitochondrial depolarization and graft injury after transplantation of fatty livers. Free Radic Biol Med 2012;53:250-9. [Crossref] [PubMed]
- Gong WF, Zhong JH, Lu Z, et al. Evaluation of liver regeneration and post-hepatectomy liver failure after hemihepatectomy in patients with hepatocellular carcinoma. Biosci Rep 2019;39:BSR20190088. [Crossref] [PubMed]
- Wang JL, Ding HR, Pan CY, et al. Mesenchymal stem cells ameliorate lipid metabolism through reducing mitochondrial damage of hepatocytes in the treatment of post-hepatectomy liver failure. Cell Death Dis 2021;12:111. [Crossref] [PubMed]
- Helling TS. Liver failure following partial hepatectomy. HPB (Oxford) 2006;8:165-74. [Crossref] [PubMed]
- Khiati S, Baechler SA, Factor VM, et al. Lack of mitochondrial topoisomerase I (TOP1mt) impairs liver regeneration. Proc Natl Acad Sci U S A 2015;112:11282-7. [Crossref] [PubMed]
- Rehman H, Liu Q, Krishnasamy Y, et al. The mitochondria-targeted antioxidant MitoQ attenuates liver fibrosis in mice. Int J Physiol Pathophysiol Pharmacol 2016;8:14-27. [PubMed]
- Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell 2012;148:1145-59. [Crossref] [PubMed]
- Li J, Miao B, Wang S, et al. Hiplot: a comprehensive and easy-to-use web service for boosting publication-ready biomedical data visualization. Brief Bioinform 2022;23:bbac261. [Crossref] [PubMed]
- Sherman BT, Hao M, Qiu J, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res 2022;50:W216-21. [Crossref] [PubMed]
- Szklarczyk D, Kirsch R, Koutrouli M, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res 2023;51:D638-46. [Crossref] [PubMed]
- van den Broek MA, Olde Damink SW, Dejong CH, et al. Liver failure after partial hepatic resection: definition, pathophysiology, risk factors and treatment. Liver Int 2008;28:767-80. [Crossref] [PubMed]
- Li H, Ge X, Pan K, et al. The Predictive Role of Tenascin-C and Cellular Communication Network Factor 3 (CCN3) in Post Hepatectomy Liver Failure in a Rat Model and 50 Patients Following Partial Hepatectomy. Med Sci Monit 2019;25:6755-66. [Crossref] [PubMed]
- Kuramitsu K, Gallo D, Yoon M, et al. Carbon monoxide enhances early liver regeneration in mice after hepatectomy. Hepatology 2011;53:2016-26. [Crossref] [PubMed]
- Liu W, Shi Y, Cheng T, et al. Weighted Gene Coexpression Network Analysis in Mouse Livers following Ischemia-Reperfusion and Extensive Hepatectomy. Evid Based Complement Alternat Med 2021;2021:3897715. [Crossref] [PubMed]
- Ou-Yang Q, Lin XM, Zhu YJ, et al. Distinct role of nuclear receptor corepressor 1 regulated de novo fatty acids synthesis in liver regeneration and hepatocarcinogenesis in mice. Hepatology 2018;67:1071-87. [Crossref] [PubMed]
- Gonzales E, Julien B, Serrière-Lanneau V, et al. ATP release after partial hepatectomy regulates liver regeneration in the rat. J Hepatol 2010;52:54-62. [Crossref] [PubMed]
- Flis DJ, Olek RA, Kaczor JJ, et al. Exercise-Induced Changes in Caveolin-1, Depletion of Mitochondrial Cholesterol, and the Inhibition of Mitochondrial Swelling in Rat Skeletal Muscle but Not in the Liver. Oxid Med Cell Longev 2016;2016:3620929. [Crossref] [PubMed]
- Cadena LR, Gahura O, Panicucci B, et al. Mitochondrial Contact Site and Cristae Organization System and F1FO-ATP Synthase Crosstalk Is a Fundamental Property of Mitochondrial Cristae. mSphere 2021;6:e0032721. Erratum in mSphere 2022;7:e0018922. [Crossref] [PubMed]
- Shi Y, Liu W, Wang K. Relationship between mitochondrial protein Mic60/OPA1 and energy supply during liver regeneration in mice. DangDaiYiXue 2021;27:154-157.
- Dong T, Zhang ZQ, Sun LH, et al. Mic60 is essential to maintain mitochondrial integrity and to prevent encephalomyopathy. Brain Pathol 2023;33:e13157. [Crossref] [PubMed]
- Vercellino I, Sazanov LA. The assembly, regulation and function of the mitochondrial respiratory chain. Nat Rev Mol Cell Biol 2022;23:141-61. [Crossref] [PubMed]
- Fiedorczuk K, Sazanov LA. Mammalian Mitochondrial Complex I Structure and Disease-Causing Mutations. Trends Cell Biol 2018;28:835-67. [Crossref] [PubMed]
- Formosa LE, Dibley MG, Stroud DA, et al. Building a complex complex: Assembly of mitochondrial respiratory chain complex I. Semin Cell Dev Biol 2018;76:154-62. [Crossref] [PubMed]
- Bahadir A, Erduran E, Değer O, et al. Augmented mitochondrial cytochrome c oxidase activity in children with iron deficiency: a tandem between iron and copper? Arch Med Sci 2018;14:151-6. [Crossref] [PubMed]
- Rak M, Bénit P, Chrétien D, et al. Mitochondrial cytochrome c oxidase deficiency. Clin Sci (Lond) 2016;130:393-407. [Crossref] [PubMed]
- Nesci S, Trombetti F, Ventrella V, et al. Opposite rotation directions in the synthesis and hydrolysis of ATP by the ATP synthase: hints from a subunit asymmetry. J Membr Biol 2015;248:163-9. [Crossref] [PubMed]
- Papa S, Martino PL, Capitanio G, et al. The oxidative phosphorylation system in mammalian mitochondria. Adv Exp Med Biol 2012;942:3-37. [Crossref] [PubMed]
- Chenna S, Koopman WJH, Prehn JHM, et al. Mechanisms and mathematical modeling of ROS production by the mitochondrial electron transport chain. Am J Physiol Cell Physiol 2022;323:C69-83. [Crossref] [PubMed]
- Lee JH, Park A, Oh KJ, et al. The Role of Adipose Tissue Mitochondria: Regulation of Mitochondrial Function for the Treatment of Metabolic Diseases. Int J Mol Sci 2019;20:4924. [Crossref] [PubMed]
- Xie TR, Liu CF, Kang JS. Dye-based mito-thermometry and its application in thermogenesis of brown adipocytes. Biophys Rep 2017;3:85-91. [Crossref] [PubMed]
- Lynes MD, Kodani SD, Tseng YH. Lipokines and Thermogenesis. Endocrinology 2019;160:2314-25. [Crossref] [PubMed]
- Friedman SL, Neuschwander-Tetri BA, Rinella M, et al. Mechanisms of NAFLD development and therapeutic strategies. Nat Med 2018;24:908-22. [Crossref] [PubMed]
- Jonckheere AI, Renkema GH, Bras M, et al. A complex V ATP5A1 defect causes fatal neonatal mitochondrial encephalopathy. Brain 2013;136:1544-54. [Crossref] [PubMed]
- Zhang G, Zhong J, Lin L, et al. Loss of ATP5A1 enhances proliferation and predicts poor prognosis of colon adenocarcinoma. Pathol Res Pract 2022;230:153679. [Crossref] [PubMed]
- Liu X, Li M, Chen Z, et al. Mitochondrial calpain-1 activates NLRP3 inflammasome by cleaving ATP5A1 and inducing mitochondrial ROS in CVB3-induced myocarditis. Basic Res Cardiol 2022;117:40. [Crossref] [PubMed]
- Zuo J, Lei M, Wen M, et al. Overexpression of ATP5b promotes cell proliferation in asthma. Mol Med Rep 2017;16:6946-52. [Crossref] [PubMed]
- Wang X, Chang X, He C, et al. ATP5B promotes the metastasis and growth of gastric cancer by activating the FAK/AKT/MMP2 pathway. FASEB J 2021;35:e20649. [Crossref] [PubMed]
- Jin P, Jiang J, Zhou L, et al. Disrupting metformin adaptation of liver cancer cells by targeting the TOMM34/ATP5B axis. EMBO Mol Med 2022;14:e16082. [Crossref] [PubMed]
- Yang W, Zhang K, Zhang Z, et al. Claudin-10 overexpression suppresses human clear cell renal cell carcinoma growth and metastasis by regulating ATP5O and causing mitochondrial dysfunction. Int J Biol Sci 2022;18:2329-44. [Crossref] [PubMed]
- Sato A, Takagi K, Miki Y, et al. Cytochrome c1 as a favorable prognostic marker in estrogen receptor-positive breast carcinoma. Histol Histopathol 2019;34:1365-75. [PubMed]
- Ramasubramanian A, Arumugam P, Ramani P, et al. Identification of Novel Cytochrome C1 (CYC1) Gene Expression in Oral Squamous Cell Carcinoma- An Evaluative Study. Ann Maxillofac Surg 2022;12:144-50. [Crossref] [PubMed]
- Chishiki M, Takagi K, Sato A, et al. Cytochrome c1 in ductal carcinoma in situ of breast associated with proliferation and comedo necrosis. Cancer Sci 2017;108:1510-9. [Crossref] [PubMed]
- Van der Schueren B, Vangoitsenhoven R, Geeraert B, et al. Low cytochrome oxidase 4I1 links mitochondrial dysfunction to obesity and type 2 diabetes in humans and mice. Int J Obes (Lond) 2015;39:1254-63. [Crossref] [PubMed]
- Binder NK, Sheedy JR, Hannan NJ, et al. Male obesity is associated with changed spermatozoa Cox4i1 mRNA level and altered seminal vesicle fluid composition in a mouse model. Mol Hum Reprod 2015;21:424-34. [Crossref] [PubMed]
- Abu-Libdeh B, Douiev L, Amro S, et al. Mutation in the COX4I1 gene is associated with short stature, poor weight gain and increased chromosomal breaks, simulating Fanconi anemia. Eur J Hum Genet 2017;25:1142-6. [Crossref] [PubMed]
- Shukla KK, Kwon WS, Rahman MS, et al. Nutlin-3a decreases male fertility via UQCRC2. PLoS One 2013;8:e76959. [Crossref] [PubMed]
- Bansept C, Gaignard P, Lebigot E, et al. UQCRC2-related mitochondrial complex III deficiency, about 7 patients. Mitochondrion 2023;68:138-44. [Crossref] [PubMed]
- Shang Y, Zhang F, Li D, et al. Overexpression of UQCRC2 is correlated with tumor progression and poor prognosis in colorectal cancer. Pathol Res Pract 2018;214:1613-20. [Crossref] [PubMed]


