Next generation sequencing identifies ‘interactome’ signatures in relapsed and refractory metastatic colorectal cancer
Introduction
Colorectal cancer (CRC) remains a significant healthcare issue representing the 4th most frequently diagnosed cancer and the second leading cause of cancer mortality in the United States. In 2015 alone, nearly 133,000 estimated new cases of CRC were diagnosed, reflecting eight percent of all new cancer cases. An estimated 49,700 cancer deaths from CRC is predicted in 2015 (1). While the burden of disease is undeniably evident, therapeutic progress is being made in the treatment of CRC. The Center for Disease Control (CDC) has reported that between 2008 and 2011, the incidence of CRC has decreased at a rate of 4% per year. Morality rate has also decreased by nearly 35% in 1990–2007 and in 2011, down by 47% (1,2). This improvement in incidence and overall mortality rate is a function of at least three known improvements in the approach and management of CRC. First, a concerted effort for improved cancer prevention across the nation. Second, increased community awareness through active colorectal screening protocols resulting in prompt diagnosis and referral. Third, a recent surge of FDA approved novel chemotherapeutic and biologic treatment modalities have increased the oncologists’ therapeutic repertoire to manage this disease. Metastatic colorectal cancer (mCRC) represents 20% of newly diagnosed CRC and through a multidisciplinary approach with the utilization of systemic backbone chemotherapy, biologic, targeted therapy and evolving surgical and radiologic interventions, the overall survival (OS) has improved to greater than thirty months.
In regards to the management of mCRC, expanded RAS mutation testing to include KRAS and NRAS codons 12 and 13 (exon 2), 59 and 61 (exon 3), and 117 and 146 (exon 4) is a predictive biomarker and guides therapeutic options (3,4). BRAF V600E mutation has also been validated to be a prognostic biomarker identifying a unique cohort of patients (pts) who will have a more aggressive clinical course (5). Recently, an exciting relationship between microsatellite instability high (MSI-H) and an impressive response to immune checkpoint blockade with anti-PD1 therapy in mCRC pts has been established (6). This reflects the hypothesis that MSI-H CRC harbors thousands of somatic mutations generating neo-antigens that are likely susceptible to immune checkpoint blockade via anti-PD1 therapy. These data reaffirm that genomics continues to emerge as a critical component in the management of advanced CRC pts. The seminal work conducted by the Human Genome Project has paved the way for subsequent Cancer Genome Atlas investigations that surveyed the specific genetic make-up of numerous malignancies in order to promote a deeper molecular understanding of cancer and provided potential clinical insights (7).
Next generation sequencing (NGS) has now become readily available and markedly affordable facilitating widespread clinical accessibility of ‘personalized’ genetic aberrations in advanced cancer pts. Although we can all agree NGS offers a wealth of genomic information, its implications in ensuring true precision has not been completely defined. Therefore, ‘precision medicine’ in oncology today should be defined as an evolving analysis with goals of correlating patient specific genetic data with prognosis, predicting response and resistance to therapy to guide treatment decisions for an individual patient. Considering that tumors consist of distinct molecular dynamics, comprised of DNA mutations, chromosomal abnormalities, gene and epigenetic expressions and proteomics reflects vast tumor heterogeneity. The new taxonomy for cancer is clearly here to stay where histology alone is no longer sufficient. Therefore, the incorporation of large scale genomic sequencing is appropriately redefining how we categorize cancer today and for the medical oncologist there is a need to accurately elucidate what role it will play in decision making for precision medicine.
A large global consortium has characterized CRC into four distinct ‘consensus molecular subtypes’ reflecting a new era of oncologic taxonomy for CRC (8). This paradigm shifting approach to CRC will undoubtedly translate into innovative ‘personalized’ clinical trial design that investigates potentially actionable genomic variants while simultaneously seeking biomarker validation in carefully pre-selected cohorts. We believe that as medical oncologists investigating specific mutational profiles can be correlated with clinical phenotypes however consideration of the entire ‘molecular signature’ to promote translational and functional studies is a high priority. These studies can serve as a means to better understand the biology of CRC, identify novel targets, unveil anticipated responses and acquired mechanisms of resistance to targeted therapy. Risk-based stratification of pts promotes innovative genomic driven prospective clinical trials. The landscape of clinical trials has already dramatically shifted to a genomic focus for both ASCO and NCI (9,10). We sought to further investigate the molecular signatures of our cohort of relapsed and refractory mCRC pts through the use of NGS platforms to evaluate pathway-network analysis to highlight how NGS data can be incorporated into the clinic today. We hypothesized that relapsed and refractory mCRC pts possess unique mutations that drive distinct aberrant pathways-networks that help understand pathobiology and generate novel therapies to be evaluated in early phase clinical trials.
Methods
Thirty-two relapsed and refractory mCRC pts (N=32) were profiled by commercially available NGS platforms: Caris Molecular Intelligence (IHC, FISH/CISH, NGS) and/or Foundation One (NGS, copy number). Tumor samples were formalin-fixed, paraffin-embedded (FFPE) blocks from diagnostic biopsies and/or metastatic sites. Each mutational profile reported histology, primary and/or metastatic site, biopsy location, gene mutation (G-site/G-mutation; P-site/P-mutation), domain, topology, and mutation count/gene. All patient information was de-identified. We then utilized web-based bioinformatics tools (Enrichr/Reactome) to carefully analyze each mutational profile identifying common and/or novel signaling as well as potential feedback loop pathway-networks, representing individual ‘mutational signatures’ (11-13).
Enrichr is a free web-based gene signature search engine that contains over 90,000 annotated signatures allowing for downstream signal and functional analytics. It encompasses over 30 gene-set libraries and various interactive visualization approaches to informatively display enrichment results via JavaScript library Data Driven Documents (D3) (12,13). This tool provides a visualization summary of known pathways based on a collective gene function list. Reactome is a free, open-source, curated and peer reviewed pathway database available for online use. This bioinformatics database provides a mechanism for biologic interpretation and visualization models for network pathway analysis. Pathway network interactions and annotations are authored by expert biologists in the field as well as cross-referenced to numerous bioinformatics databases, such as NCBI Gene, Ensembl and UniProt databases, the UCSC and HapMap Genome Browsers, the KEGG Compound and ChEBI small molecule databases, PubMed, and Gene Ontology (11,13). Through the use of Reactome and Enrichr, we were able to systematically visualize anticipated pathway networks based upon patient specific molecular profiles. These bioinformatics tools allow for prompt visualization and analysis of involved signaling pathway networks fostering further hypothesis generation for biomarker development.
Results
Mutational frequencies in relapsed and refractory mCRC
All mCRC pts included in this analysis had progressed on fluoropyrimidines, oxaliplatin, irinotecan, bevacizumab, cetuximab or panitumumab. Adenocarcinoma was the most common histology followed by squamous cell carcinoma (colon N=29; rectal N=3). In our cohort of thirty-two relapsed and refractory mCRC pts, we identified the following oncogenes and genetic aberrations in highest frequency—TP53, APC, KRAS, PIK3CA, BRAF, SMAD4, SPTA1, FAT1, PDGFRA, ATM, ROS1, ALK, CDKN2A, FBXW7, TGFBR2, NOTCH1 and HER3 (Figure 1, Table 1). Most patients had on average >5 unique gene mutations. Five patients were identified with MSI-H. High mutational burden was not predictive for PD-1 (n=5) or PD-L1 (n=1) positivity (Figure 1).
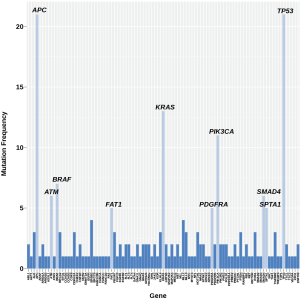
Full table
The ‘interactome’ in mCRC
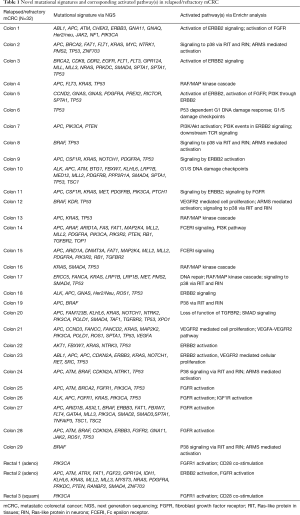
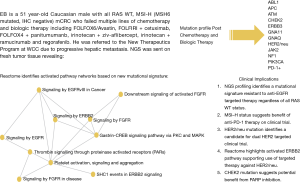
There are no simple genotype-phenotype relationships in mCRC as highlighted by the BRAF mutation status. In refractory melanoma vemurafenib showed a 78% response rate but in mCRC the response rate was 5% (14,15). Thus, a dominant oncogene mutation does not imply oncogenic dependence, as pathway-network status is distinct to each tumor type. Hence, biologic context referred to here as the ‘interactome’ i.e., network of activated pathways and feedback loops, confound isolated targeting of a specific genetic aberration. Enrichr and Reactome tools were utilized to interrogate signaling pathways concomitantly represented in our cohort of mCRC pts based upon their specific mutational profiles (Figure 2). Collectively the interactome generated and identified a map of signaling nodes that predict for activation of four interacting pathways in mCRC: (I) Erb-B2 receptor kinase 2 (HER2); (II) fibroblast growth factor receptor 1 (FGFR1); (III) p38 activation through BRAF-MEK cascade via Ras-like protein in tissues (RIT) and Ras-like protein in neurons (RIN) GTPases; (IV) ARMS-mediated activation of MAPK cascade; and (V) VEGFR2 (Figure 2). In addition, there were eight cases with DNA-repair defects with mutations in ATM, CHEK2, FANCA and/or BRCA2 that may warrant further investigation with PARP inhibition (Table 1) (16,17). Whether the development of somatic mutations in DNA repair genes are secondary to heavy pretreatment with chemotherapy would need to be investigated, thereby identifying an expected tumor evolution signature that could potentially be exploited with PARP inhibition in refractory mCRC. Tumor evolution was further appreciated within our cohort of patients when NGS data from new liver metastasis was obtained in a patient with expanded RAS wild-type mCRC heavily pretreated with chemotherapy, anti-VEGF therapy and anti-EGFR therapy that subsequently developed coexistent KRAS (Q61L) plus NRAS (Q61H) mutations (Table 1). To highlight both the importance and complexity of the “interactome” as a critical component for choosing next best steps in therapy, we also describe a case of heavily pre-treated relapsed and refractory RAS wild type, MSI-H mCRC (Figure 3).
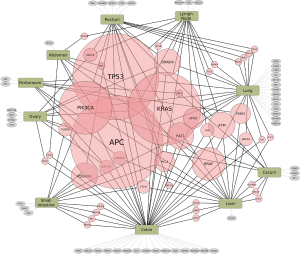
HER2 activation in RAS wild type and mutated CRC
In our cohort of relapsed and refractory mCRC of all RAS wild type pts previously treated with anti-EGFR therapy and RAS mutant pts treated with cytotoxic chemotherapy plus VEGF inhibition, the most common activated signaling pathway was ErbB2/Her2neu. Our data imply that HER2 is an important mechanism of resistance to frontline therapies in RAS wild type and mutated mCRC. Overexpression and/or activation of HER2 as well as significant crosstalk between other ErbB family members and subsequent downstream signaling is a suspected resistance mechanism to anti-EGFR therapy for all RAS wild type mCRC. Targeting of HER2 overexpressing mCRC is being investigated in the HER2 Amplification for Colorectal Cancer Enhanced Stratification (HERACLES) phase II study, initially begun in Italy in August 2012 to enroll 27 KRAS wild type refractory mCRC pts overexpressing HER2 to receive trastuzumab plus lapatinib, with a subsequent 27 pts to receive trastuzumab plus pertuzumab (18). In RAS mutant mCRC pts we also identified an over-activation of the ErbB2 pathway which suggests HER2 is a bystander or an active oncogenic partner by activating the PI3K/AKT/mTor pathway in addition to amplified RAS/MAPK signaling. Targeting HER2 in this setting (e.g., anti-HER2 plus lapatinib) may provide further clues to the validity of this approach.
HER3 mutations in mCRC
We identified five HER3 mutations in our cohort of relapsed and refractory RAS wild type and mutated mCRC pts. They include: one patient with three mutations on exon 6/A232V (pathogenic), exon 20/Q809R (pathogenic) and exon 23/E928G (variant of unknown significance-VUS); a second patient with exon 21/H828R (VUS) and a third patient with exon 2/T68M (VUS). HER3 is a kinase depleted co-receptor that is successful in downstream signaling after forming heterodimers with other ErbB family members in order to bind its ligand heregulin (19) which activates the PI3K pathway (20). Preclinical work has revealed that HER3 overexpression by IHC is a prognostic marker for inferior survival in CRC. A phase I study in refractory cancer pts investigating MEHD7945A, a human IgG1 antibody with dual binding to HER3 and EGFR, found 4 of 12 CRC pts developed stable disease for >8 weeks. Currently a phase II study is actively recruiting pts with refractory KRAS wild type mCRC to treatment arms with FOLFIRI plus MEHD7945A versus FOLFIRI plus cetuximab (NCT01652482). Dual targeting of HER2/HER3 plus PI3K/AKT/mTor pathway is an approach for these pts. Previous work established PIK3CA mutations as a prognostic biomarker in CRC considering that aspirin use in PIK3CA mutated CRC results in longer survival compared to wild-type PIK3CA CRC, supporting secondary prevention with aspirin therapy (21). Due to its effect via PI3K/AKT/mTOR pathway on cell growth and survival and representing a compensatory feedback resistance pathway in response to anti-EGFR therapy, this represents a key target in mCRC. Early phase clinical trials combining dual PI3K/mTOR inhibition and/or cytotoxic chemotherapy with anti-EGFR therapy are underway to ascertain the potential outcome of targeting this alteration in mCRC (22,23).
Implications for FGFR Pathway in mCRC
FGFR1, FGFR2, and FGF23 aberrations were identified by NGS analysis in wild type and mutated KRAS pts with mCRC. Brivanib alaninate, the L-alanine ester prodrug of brivanib, an oral, potent and small molecule inhibitor of VEGFR/FGFR tyrosine kinases (24) was tested in a phase III trial of 750 pts randomized to cetuximab plus brivanib alaninate versus cetuximab plus placebo in pts with metastatic chemotherapy refractory wild type KRAS mCRC (CO.20) (25). Pts enrolled on study received at least one prior line of therapy with no prior exposure to anti-EGFR therapy. The cetuximab plus brivanib had a statistically significant progression free survival (PFS) (5.0 vs. 3.4 months; HR, 0.72; P<0.001) and a partial response rate of 13.6% vs. 7.2% favoring the combination arm (P=0.004). However, there was no statistically significant improvement in OS (8.8 vs. 8.1 months; P=0.12) with worsening grade 3 non-hematologic toxicities with cetuximab plus brivanib. Although a ‘negative’ study, there were interesting findings. For example, the discordance between PFS and OS and the poor quality of life metrics likely has less to do with FGFR targeted therapy but more reflective of an unselected patient population entering a targeted clinical trial. To date, there are no validated predictive biomarkers for the appropriate selection of patients who will respond to FGFR inhibition in mCRC. Our data reveal a cohort of mCRC pts with FGFR1, FGFR2 and FGF23 mutations with associated FGFR pathway activation. Considering the PFS results in CO.20, there is indeed a cohort of pts within the total cohort that had clear response to therapy with FGFR inhibition, however these pts were not preselected based on a validated biomarker. Retrospective analysis of the CO.20 trial may serve to highlight and validate a biomarker of response in mCRC pts with the utilization of IHC, FISH and NGS to correlate with response. Further investigation using NGS and concomitant pathway analysis to stratify mCRC pts who would benefit most from FGFR inhibition is warranted.
Anti-angiogenic signature in mCRC
In our cohort of relapsed and refractory RAS wild type and mutated mCRC pts, we identified PDGFRA as one of the most common mutation frequencies (Figure 1) and in conjunction with pathway analysis we noted 3 pts with PDGFRA mutations and corresponding FGFR pathway activation (Table 1). We identified 2 pts with VEGFA and KDR (VEGFR2) mutations with corresponding VEGFR2 pathway activation (Table 1). The CORRECT trial led to the approval of regorafenib in mCRC pts with a 1.4-month OS (HR 0.77, P=0.0052) compared to placebo (24,26). Pts were not selected based on an ‘angiogenic’ biomarker to determine clinical response to regorafenib which may have shown benefit if the biomarker was predictive of response. Correlative studies in the CORRECT trial investigated plasma angiogenesis proteins as potential biomarkers. Elevated level of TIE-1 was found to be associated with improved outcomes with regorafenib in terms of OS (27). Also, previous genotypic work correlated regorafenib treatment outcomes with single nucleotide polymorphisms in VEGF and VEGFR. The presence of VEGF-A rs2010963 was independently correlated with PFS and OS (HR: 0.49, 95% CI, 0.33–0.81 and HR: 0.52, 95% CI, 0.34–0.99), representing promise as a potential biomarker (26). Using NGS data to identify pts angiogenic targets (e.g., PDGFRA, VEGFA, KDR, FLT1, FLT4) and CMS4 (mesenchymal, stromal infiltration and increased angiogenesis) may provide insights in the regorafenib patient cohort in the CORRECT trial. Pts who develop toxicities to regorafenib do so in the first few weeks of initiating treatment, one could hypothesize that early toxicities manifest in tumors that are not biologically dependent on regorafenib. Identifying a selected population for TKI therapy may be clinically meaningful with an improved quality of life.
Discussion
Molecular subtyping coupled to a deeper understanding of cancer biology is clearly shifting the paradigm of cancer treatments. Biomarker approach to therapy has had some success as evident in metastatic breast cancer, lung cancer, gastric cancer and colon cancer enabling careful preselection of pts that benefit from targeted therapies (28-31). In 2015, elegant data reveals that MSI alone identifies pts in both CRC and non-CRC cohorts that may benefit from checkpoint blockade with anti-PD-1 monoclonal antibody therapy (6). Therefore, implementing the use of NGS in order to ascertain key genetic characteristics of tumors is instructive. Current understanding mandates a multimodality approach involving cytotoxic chemotherapy, targeted therapy and immune checkpoint therapy. However, considering a more favorable toxicity profile, numerous clinical trials have been designed to compare targeted molecular or immune therapy to standard of care cytotoxic therapy (32,33). Despite current advances (3,30), new therapies targeting angiogenesis (e.g., ziv-aflibercept, ramucirumab, regorafenib) have not moved the field significantly (26,34,35). Cytotoxic chemotherapy continues to add modestly in relapsed and refractory mCRC with the recent FDA approval of TAS-102, a novel oral combination of trifluridine, a thymidine-based nucleic acid analogue with a thymidine phosphorylase inhibitor, tipiracil (36).
Although in the relapsed and refractory setting of mCRC, NGS is being performed in academia and in community oncology clinics, a clear step wise systematic approach to when to order and how to interpret data is not established. We utilized NGS to test the hypothesis that dominant oncogene mutations do not always equate to the phenotype and an understanding of the underlying interactome in each patient provides clues to choosing the next best therapy, hopefully within a clinical trial setting. We highlight that the ‘interactome’ derived from NGS represent a meaningful tool to: (I) evaluate functional investigation of novel candidate molecular targets in the laboratory; (II) predict response and/or resistance to drug therapy; (III) guide rational drug combinations in relapsed and refractory mCRC pts.
Recently, four consensus molecular subtypes (CMS) for CRC was identified by an international consortium (8) based on six independent classification systems from 18 CRC data sets, including TCGA representing 4,151 patients. These include: CMS1 (14%), defined as MSI, CpG island methylator phenotype (CIMP) high, hypermutated, associated with BRAF mutations and signaling pathways significant for immune infiltration and activation; CMS2 (37%), defined as canonical, with somatic copy number alterations (SCNA) and WNT and MYC pathway activation; CMS3 (13%), defined as metabolic with mixed MSI status, SCNA low, CIMP low, associated with KRAS mutations and pathways responsible for metabolic dysregulation; CMS4 (23%), SCNA high with pathways responsible for stromal infiltration, TGF-β activation, and angiogenesis. A mixed feature group reflected the remaining 13% of tumors was postulated to represent a transition phenotype (8). The data also reveals that aside from the association of BRAF mutations in CMS1 and KRAS mutations in CMS3, there was no isolated event or specific genetic alteration limited to each subtype. The development of EGFR ectodomain S492R mutations in KRAS WT pts treated with cetuximab therapy represents an example of this concept producing acquired resistance to biologic therapy (37). We present a redefined approach to mCRC pts using NGS plus pathway-network analysis within the CMS categories to test targeting aberrant biology in clinical trials (Figure 4).
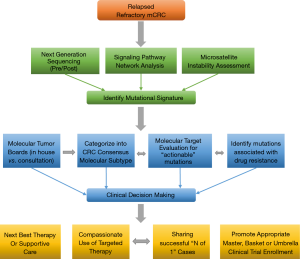
Incorporation of CMS and NGS plus pathway-network analysis may better guide the management of pts with advanced mCRC. Our study represents a novel systematic approach to incorporating ‘mutational signatures’ into an ‘interactome’ in clinical decision making. This approach will aid in the identification of new targets that warrant further large scale prospective investigation, highlight potential biomarkers of response and resistance. In this paper we illustrate that dominant oncogene mutations do not always equate with oncogenic dependence and understanding cross-talk between activated signaling pathways and feedback loops in heavily pretreated relapsed and refractory mCRC pts is key to successful drug development. Our work is hypothesis generating for identifying novel targets and corresponding networks. NGS enhances elucidation of tumor clonal evolution and pathway-network analysis elaborating new and anticipated drug resistance mechanisms. The PROSPECT-C, reported that obtaining circulating tumor DNA of a patient with mCRC while on cetuximab therapy revealed a compensatory rise and fall in mutant RAS clones in response to the presence or absence of anti-EGFR therapy (38). As reported earlier within our cohort of pts, NGS data from new liver metastasis was obtained in a patient with RAS wild-type mCRC heavily pretreated with chemotherapy, anti-VEGF therapy and anti-EGFR therapy that subsequently developed coexistent KRAS (Q61L) plus NRAS (Q61H) mutations. This showcases the “real-time” insight obtaining ‘pre/post’ NGS data, provides to document evidence of tumor evolution and facilitates a personalized approach to next best therapy in acquired genomic changes.
We propose an approach regarding the utilization of genomic data in the management of pts with mCRC (Figure 4). This approach represents an organized means to incorporate NGS and pathway-networks of each patient with mCRC furthering new research hypothesis, novel clinical trial design and enrollment using “N of 1” for reporting therapeutic success. This approach may result in a deeper understanding of the complex genomic landscape in advanced mCRC. Our approach (Figure 4) may aid in validation of this technology as a critical clinical decision making tool for advanced mCRC.
Conclusions
Expanded RAS mutational status in CRC is a clinically validated but very limited biomarker that guides current therapeutic modalities. Emerging data indicate that BRAF mutation status is prognostic and predictive for response to anti-EGFR therapy (39) but is also limited. Although the use of NGS in relapsed and refractory mCRC is widespread in clinical practice, guidelines reflecting systematic incorporation of genomic data into daily patient care is in its infancy. Our experience in the utilization of NGS plus bioinformatics tools to identify signaling pathways representing a distinctive ‘mutational signature’ or ‘interactome’ for individual mCRC pts may allow for a tailored approach to therapy. The mutational signatures of thirty-two pts with relapsed and refractory mCRC identified HER2, HER3, PDGFRA, ATM, ALK, ROS1, CDKN2A, and PIK3CA as potential targets of focus and HER2 and FGFR as unique activated feedback-escape pathways of interest for rational drug development. A focus on the ‘interactome’ highlights the biologic context of each patient’s mutational profile and represents a unique approach to laboratory investigation, enhancing drug development, directing next best therapy and appropriately triaging patients to enrollment on genomic clinical trials.
Acknowledgements
We wish to thank The West Cancer Center and University of Tennessee Health Science Center (UTHSC) for support. B Johnson received funding for this project from a UTHSC research grant.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: This study was approved by the UTHSC Institutional Review Board under exempt status.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015;65:5-29. [Crossref] [PubMed]
- Henley SJ, Singh SD, King J, et al. Invasive cancer incidence and survival--United States, 2011. MMWR Morb Mortal Wkly Rep 2015;64:237-42. [Crossref] [PubMed]
- Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 2008;26:1626-34. [Crossref] [PubMed]
- Sorich MJ, Wiese MD, Rowland A, et al. Extended RAS mutations and anti-EGFR monoclonal antibody survival benefit in metastatic colorectal cancer: a meta-analysis of randomized, controlled trials. Ann Oncol 2015;26:13-21. [Crossref] [PubMed]
- Van Cutsem E, Köhne CH, Láng I, et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol 2011;29:2011-9. [Crossref] [PubMed]
- Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330-7. [Crossref] [PubMed]
- Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015;21:1350-6. [Crossref] [PubMed]
- TAPUR. Targeted Agent and Profiling Utilization Registration Study. Available online: http://www.tapur.org. Accessed Dec 21, 2015.
- Conley BA, Doroshow JH. Molecular analysis for therapy choice: NCI MATCH. Semin Oncol 2014;41:297-9. [Crossref] [PubMed]
- Croft D, Mundo AF, Haw R, et al. The Reactome pathway knowledgebase. Nucleic Acids Res 2014;42:D472-7. [Crossref] [PubMed]
- Chen EY, Tan CM, Kou Y, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 2013;14:128. [Crossref] [PubMed]
- The Reactome pathway knowledgebase’ (Available online: ) (Reactome, 2015) and Enrichr (Available online: ). Accessed Dec 21, 2015.http://www.reactome.org
- Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med 2010;363:809-19. [Crossref] [PubMed]
- Kopetz S, Desai J, Chan E, et al. Phase II Pilot Study of Vemurafenib in Patients With Metastatic BRAF-Mutated Colorectal Cancer. J Clin Oncol 2015;33:4032-8. [Crossref] [PubMed]
- Fong PC, Yap TA, Boss DS, et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol 2010;28:2512-9. [Crossref] [PubMed]
- Mateo J, Carreira S, Sandhu S, et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N Engl J Med 2015;373:1697-708. [Crossref] [PubMed]
- Marsoni S, Bertotti A, Sartore-Bianchi A, et al. Dual anti-HER2 treatment of patients with HER2-positive metastatic colorectal cancer: The HERACLES trial (HER2 Amplification for Colo-rectaL Cancer Enhanced Stratification). J Clin Oncol 2013;31:abstr TPS3648.
- Jones JT, Akita RW, Sliwkowski MX. Binding specificities and affinities of egf domains for ErbB receptors. FEBS Lett 1999;447:227-31. [Crossref] [PubMed]
- Soltoff SP, Carraway KL 3rd, Prigent SA, et al. ErbB3 is involved in activation of phosphatidylinositol 3-kinase by epidermal growth factor. Mol Cell Biol 1994;14:3550-8. [Crossref] [PubMed]
- Liao X, Lochhead P, Nishihara R, et al. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med 2012;367:1596-606. [Crossref] [PubMed]
- Mallon R, Feldberg LR, Lucas J, et al. Antitumor efficacy of PKI-587, a highly potent dual PI3K/mTOR kinase inhibitor. Clin Cancer Res 2011;17:3193-203. [Crossref] [PubMed]
- Tabernero J, Brega N, Davis C, et al. A randomized phase II study (B2151005) of the intravenous phosphatidylinositol 3-kinase (PI3K)/mammalian target of rapamycin (mTOR) inhibitor PF-05212384 plus irinotecan versus cetuximab plus irinotecan in patients with wild-type KRAS metastatic colorectal cancer (mCRC). J Clin Oncol 2014;32:abstr TPS3649.
- Diaz-Padilla I, Siu LL. Brivanib alaninate for cancer. Expert Opin Investig Drugs 2011;20:577-86. [Crossref] [PubMed]
- Siu LL, Shapiro JD, Jonker DJ, et al. Phase III randomized, placebo-controlled study of cetuximab plus brivanib alaninate versus cetuximab plus placebo in patients with metastatic, chemotherapy-refractory, wild-type K-RAS colorectal carcinoma: the NCIC Clinical Trials Group and AGITG CO.20 Trial. J Clin Oncol 2013;31:2477-84. [Crossref] [PubMed]
- Grothey A, Van Cutsem E, Sobrero A, et al. Regorafenib monotherapy for previously treated metastatic colorectal cancer (CORRECT): an international, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet 2013;381:303-12. [Crossref] [PubMed]
- Tabernero J, Lenz HJ, Siena S, et al. Analysis of circulating DNA and protein biomarkers to predict the clinical activity of regorafenib and assess prognosis in patients with metastatic colorectal cancer: a retrospective, exploratory analysis of the CORRECT trial. Lancet Oncol 2015;16:937-48. [Crossref] [PubMed]
- Baselga J, Cortés J, Kim SB, et al. Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. N Engl J Med 2012;366:109-19. [Crossref] [PubMed]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 2010;376:687-97. [Crossref] [PubMed]
- Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 2008;359:1757-65. [Crossref] [PubMed]
- Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:123-35. [Crossref] [PubMed]
- Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:1627-39. [Crossref] [PubMed]
- Van Cutsem E, Tabernero J, Lakomy R, et al. Addition of aflibercept to fluorouracil, leucovorin, and irinotecan improves survival in a phase III randomized trial in patients with metastatic colorectal cancer previously treated with an oxaliplatin-based regimen. J Clin Oncol 2012;30:3499-506. [Crossref] [PubMed]
- Tabernero J, Yoshino T, Cohn AL, et al. Ramucirumab versus placebo in combination with second-line FOLFIRI in patients with metastatic colorectal carcinoma that progressed during or after first-line therapy with bevacizumab, oxaliplatin, and a fluoropyrimidine (RAISE): a randomised, double-blind, multicentre, phase 3 study. Lancet Oncol 2015;16:499-508. [Crossref] [PubMed]
- Mayer RJ, Van Cutsem E, Falcone A, et al. Randomized trial of TAS-102 for refractory metastatic colorectal cancer. N Engl J Med 2015;372:1909-19. [Crossref] [PubMed]
- Montagut C, Dalmases A, Bellosillo B, et al. Identification of a mutation in the extracellular domain of the Epidermal Growth Factor Receptor conferring cetuximab resistance in colorectal cancer. Nat Med 2012;18:221-3. [Crossref] [PubMed]
- Khan K, Vlachogianis G, Cunningham D, et al. Abstract 3589: Validation of the role of circulating tumor DNA (ctDNA) in tracking mechanisms of resistance to anti-EGFR monoclonal antibodies (AE-mABs): preliminary results of the PROSPECT-C prospective trial. Cancer Res 2015;75:3589. [Crossref]
- Pietrantonio F, Petrelli F, Coinu A, et al. Predictive role of BRAF mutations in patients with advanced colorectal cancer receiving cetuximab and panitumumab: a meta-analysis. Eur J Cancer 2015;51:587-94. [Crossref] [PubMed]


