
Genome-wide association study of the TP53 R249S mutation in hepatocellular carcinoma with aflatoxin B1 exposure and infection with hepatitis B virus
Introduction
Primary liver cancer is one of the most prevalent types of cancer with over 841,000 new cases recorded and causing approximately 782,000 deaths worldwide in 2018. Hepatocellular carcinoma (HCC) represents 75–85% of total liver cancer cases (1). The main risk factors for HCC are chronic infection with hepatitis B virus (HBV) or hepatitis C virus, consuming food contaminated with aflatoxin, heavy alcohol intake, obesity, smoking, and type 2 diabetes (2,3). Chronic HBV infection is associated with 53% of HCC cases worldwide (4) and an even higher proportion of cases (75%) in Asia (5). Aflatoxin B1 (AFB1) is the major carcinogenic form of aflatoxin and has been classified as a group I carcinogen in humans by the International Agency for Research on Cancer (IARC) (6). It is also a major risk factor for HCC. In 2013, 65.9% of male liver cancer-related deaths in China and 58.4% of female liver cancer-related deaths in China could be attributed to HBV infection, and an estimated 25.0% of all liver cancer-related deaths in China could be attributed to aflatoxin exposure (7).
TP53 is a tumor suppressor gene that responds to diverse cellular stress signals and regulates the expression of target genes that induce cell cycle arrest, apoptosis, senescence, DNA repair, and changes in metabolism. Mutations in the TP53 gene are the most common to occur in human malignancies (8). TP53 mutations are reportedly associated with the prognosis of multiple cancers and are biomarkers of environmental carcinogens (9). In particular, the R249S mutation in exon 7 of TP53 is a recognized molecular fingerprint of aflatoxin exposure (10-13) and is associated with poor prognosis in patients with HCC (14). Previous reports suggest that HBV and AFB1 exposure have synergistic carcinogenic effects and increase the likelihood of the TP53 R249S mutation occurring in HCC (10,11,15). Studies have shown that complex trans-activation exists between HBx and TP53, in which HBx directly inhibits TP53 activity by binding its C-terminal (16). Kew et al. (17) investigated the effects of wild-type and mutant HBx on TP53 gene, and found that HBx mutants could inhibit TP53 expression and downstream signal transduction. Recent studies have shown that overexpression of HBx C-terminal mutants in HHT4 cells (normal liver cell lines) significantly increased colony formation efficiency, while the corresponding wild-type allele significantly decreased colony formation efficiency in HHT4 cells. At the same time, TP53 R249S mutation interact with HBx mutants to regulate cell proliferation and mitochondrial stability (18). Due to high levels of both dietary exposure to AFB1 and HBV infection rates in Guangxi, the incidence of HCC is higher in this region than the collective incidence in China; the frequency of TP53 R249S mutations in Guangxi is up to 64.9% (10,19).
A genome-wide association study (GWAS) is a study that examines characteristic genetic alterations and epigenetic profiles associated with various diseases and detects the contribution of individual genetic factors to complex diseases, enabling potential targets associated with the occurrence, development, and treatment of multiple diseases to be identified (20). We conducted a GWAS using Illumina HumanExome BeadChip-12-1_A chip, to analyze genetic susceptibility to the TP53 R249S mutation and identify genetic factors that influence this mutation in the TP53 gene in HCC patients exposed to AFB1 and HBV in the Guangxi region.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/jgo-20-510).
Methods
Study participants
A total of 485 HCC patients that received hepatectomy surgery at the Department of Hepatobiliary Surgery of the First Affiliated Hospital of Guangxi Medical University between January 2001 and November 2013 were enrolled in the GWAS. An additional 270 HCC patients were recruited between December 2013 and August 2016 for the replication study. All patients were positive for hepatitis B surface antigen and HCC after receiving hepatectomies; this was confirmed by histopathology. The clinicopathological variables of the enrolled participants were obtained from their clinical records and pathological reports. These variables included age, sex, smoking status, drinking status, pathological grade, the biobehavior of their cancer, serum alpha fetoprotein (AFP) levels, hepatic cirrhosis, radical resection status, and the use of transcatheter hepatic arterial chemoembolization (TACE). Tumor status was classified according to the Barcelona Clinic Liver Cancer (BCLC) staging system (21). Child-Pugh classification was carried out as described in previous literature (22). The presence of portal vein tumor thrombus (PVTT) was determined using previously described criteria (23). Smoking status, drinking status, and radical resection status were defined according to previous literature (24).
These studies were approved by the Ethical Review Committee of the First Affiliated Hospital of Guangxi Medical University {Approval Number: 2015 [KY-E-072]}. All participants signed written informed consent. This study conformed to the provisions of the Declaration of Helsinki (as revised in 2013).
Specimens and TP53 R249S mutation detection
HCC tissue samples were collected during surgery and immediately stored at −80 °C. Total DNA was extracted using TIANamp Genomic DNA Kits [TIANGEN Biotech (Beijing) Co, Ltd., China]. DNA concentration and purity were measured using the NanoDrop2000 system (Thermo Fisher Scientific, Waltham, MA, USA). TP53 R249S mutations were detected using Sanger sequencing after amplification by polymerase chain reaction (PCR). The sequencing primers are listed in Table S1.
Immunohistochemistry
The expression of TP53 in HCC tissue samples was evaluated using immunohistochemical staining (Figure S1). Antigen retrieval was conducted using Tris-EDTA at a high temperature for 2.5 min. Tissue slices were treated with 3% hydrogen peroxide to inactivate endogenous peroxidases. After washing with PBS, the slices were incubated with mouse antibody against human TP53 (1:150) (ZSGB-BIO ORIGENE, Beijing, China) at 4 °C, followed by incubation with a secondary antibody (Dako Cytomation, Glostrup, Denmark) for 30 min at 37 °C. Finally, the tissue slices were visualized using diaminobenzidine (DAB) (Dako Cytomation) and counterstained with hematoxylin. Positive and negative controls were included for each sample. All tissue sections were reviewed and scored blindly by two experienced pathologists. In cases of disagreement, where possible, joint review consensuses were obtained. The proportion of TP53 positive cells was calculated using the following formula: number of positive cells/total number of the cells ×100%. A TP53 positive stain was defined as ≥10% of cancer cells positive for TP53 (25,26).
Genotyping and quality control in the GWAS
All samples in the GWAS were genotyped using Illumina HumanExome BeadChip-12-1_A, which includes 242,901 markers of protein-altering genetic variants. The study flow chart and the genotyping procedure are depicted in Figures S2,S3, respectively. Genotype calling was carried out using Genotyping Module v1.0 in GenomeStudio v2011.1 with an average call rate of 99.84%. A total of 50 samples (>10%) were randomly selected, and sequence analysis of candidate loci was performed using ABI PRISM 3100 (Applied Biosystems, Shanghai Sangon Biological Engineering Technology & Services Co, Ltd, Shanghai, China). The sequencing results were 100% consistent with the BeadChip-12-1_A genotyping results.
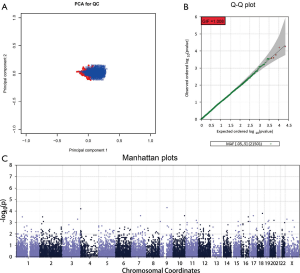
A quality control (QC) procedure was used prior to association analysis. Firstly, samples were excluded if they met any of the following criteria: (I) a genotyping rate of <95%; (II) ambiguous sex; (III) a genome-wide identity-by-descent (IBD) of >0.1875; and (IV) being an outlier in a principal components analysis (PCA) for ancestry and population stratification. Secondly, single nucleotide polymorphisms (SNPs) were removed that had (I) a genotype call rate of <95%, (II) a P value in Hardy-Weinberg equilibrium (HWE) of <1×10−6, and (III) a minor allele frequency (MAF) of <0.05. Thirdly, an analysis of population stratification by PCA was performed using the EIGENSOFT package to eliminate multi-ethnic interference. Genomic inflation factors (GIF) were used to investigate residual population stratification using MATLAB 7.0. The QC procedure was performed using Plink v1.07, R 3.0.1, and the EIGENSOFT package. After QC, the total genotyping rate in the remaining individuals was determined. This is shown in Figure S4A. The genotype failure rate and heterozygosity across all individuals are shown in Figure S4B. The PCA plot showed either no stratification or mild stratification in our study population (Figure 1A). The genomic control inflation factor was determined using a Quantile-Quantile (Q-Q) plot (Figure 1B). Ultimately, 21,501 SNPs from 459 cases were used in the GWAS following QC.
SNP selection for the replication study
Based on the presence or absence of the TP53 R249S mutation, subjects were placed into either the mutation group or the non-mutation group. Genetic differences between the two groups were explored in the GWAS. The EPACTS package was used to perform association analyses. According to the Manhattan plot produced (Figure 1C), 35 SNPs with top-ranking P values were selected. However, only 29 primer pairs could be designed for these SNPs in the replication study. The 29 SNPs used for the validation analysis are shown in Table S2, and their corresponding primers are shown in Table S3.
Genotyping and quality control in the replication study
A total of 270 patients were enrolled in the replication study. Genotyping analyses were carried out using iPLEX Gold SNP genotyping technology based on the MassARRAY array platform. The QC procedure was carried out as follows. Firstly, samples with (I) a genotyping rate of <95% and (II) a genome-wide IBD of >0.1875 were excluded. Secondly, SNPs with (I) a genotype call rate of <95%, (II) a HWE P value of <1×10−6, and (III) a MAF of <0.05 were removed. Following QC, 258 samples (80 in the mutation group and 178 in the non-mutation group) were included for analysis. The results of the replication study using the 29 SNPs are shown in Table S4, and their corresponding primers are shown in Table S5.
Quantitative real time PCR
Tumor and paired non-tumor liver tissue samples were collected for gene expression analysis from 20 HCC patients randomly selected from the independent replication study. All samples used to extract RNA were treated with RNA preservation solution and then stored in a −80° refrigerator. Candidate SNPs were selected from the GWAS, and replication analyses were then carried out to analyze potential associations between these SNPs and expression levels of their located gene. The tissue samples stored in the RNA protection solution were taken out and 10 mg of tissue was put into the centrifuge tube with Trizol reagent, and then homogenized fully with a grinder. Then the RNA extraction was carried out in accordance with Trizol’s instructions. Briefly, tissue sample Trizol lysis buffer was placed at room temperature (25 °C) for 10 minutes to further fully decompose; Then 200 µL chloroform was added and shaken vigorously for 15 seconds. Placed at room temperature (25 °C) for 5 minutes. Centrifuged at 12,000 g at 4 °C for 15 minutes. At this time, the liquid in the centrifuge tube was divided into three layers, and the RNA was mainly in the upper colorless aqueous phase. The upper colorless aqueous phase (about 500 µL) was placed into a new sterile centrifuge tube with RNase removed. Five hundred µL isopropyl alcohol was added and placed at room temperature 25 °C) for 10 minutes. The liquid was centrifuged at 12,000 g at 4 °C for 10 minutes and the supernatant was discarded. Then add 1 mL 75% ethanol for washing and discard the superfine. Let the precipitated RNA dry and then dissolve the RNA precipitate in 50 µL RNase-free water. The integrity of total RNA was determined by gel electrophoresis. cDNA reverse transcription was performed using PrimeScript™ RT Reagent Kit with gDNA Eraser (TAKARA BIO INC). Then the real-time PCR reaction was performed according to Roche FastStart Universal SYBR Green Master (ROX) kit and ABI 7500 fluorescence PCR system operating guidelines. The primers used in amplification are shown in Table S6. Quantitative real time (RT)-PCRs were conducted in triplicate on an ABI7500 RT-PCR System using FastStart Universal SYBR Green Master (Rox). The expression of genes in all samples was normalized to that of GAPDH using the 2−∆∆CT method.
Statistical analysis
Results are expressed as mean ± standard deviation (SD) for normally distributed variables and as a median for non-normal distributions. The logistic regression model was applied to analyze differences in clinicopathological factors between the TP53 R249S mutation group and the non-mutation group. Odds ratio (OR) and 95% confidence interval (CI) values were calculated using univariate analyses. Analyses of the associations between the TP53 R249S mutation and SNPs were implemented using single variant logistic score test in EPACTS package v3.2.6 (27). Q-Q and Manhattan plots were also generated. Chi-squared tests and the logistic regression model were applied to identify any associations between genetic models and gene types. Local linkage disequilibrium (LD) and recombination patterns were analyzed using LocusZoom (28). Differences in expression between tumor tissue samples and paired adjacent non-tumor tissue samples were assessed using the Student’s t test, and differences between SNP genotypes and gene expression were assessed using a non-parametric trend test. Statistical analysis was performed using SPSS v18.0 (SPSS, Chicago, IL, USA). Data were considered statistically significant when P<0.05.
Results
Baseline characteristics
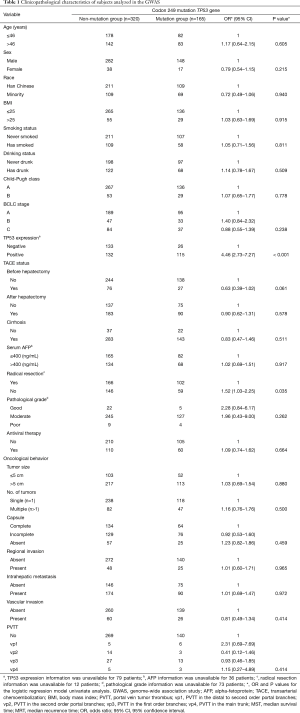
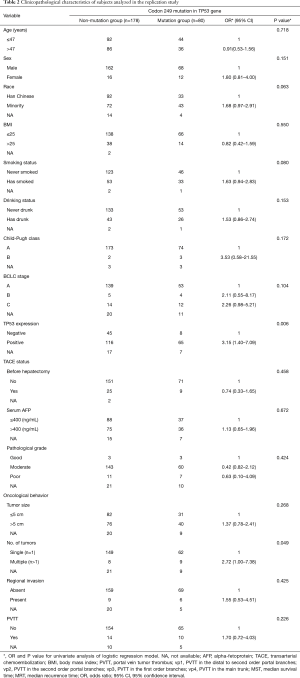
A total of 165 HCC patients with the TP53 R249S mutation were analyzed in this GWAS, as well as 320 patients without this mutation. Our replication study included 80 patients with this mutation and 178 without. When subgroups were stratified by the TP53 R249S mutation, no difference in clinicopathological parameters was found between these subgroups in either the GWAS or the replication study. These parameters included age, sex, race, BMI, smoking status, drinking status, Child-Pugh score, BCLC stage, TACE status, cirrhosis, serum AFP levels, pathological grade, antiviral therapy, tumor size, capsule status, regional invasion, intrahepatic metastasis, vascular invasion, and PVTT. There was, however, a significant difference in tumor number found in the replication study between these subgroups (HR =2.72, 95% CI: 1.00–7.38, P<0.049) (Table 1 and Table 2). TP53 expression status was found to be significantly different between the mutation group and the non-mutation group in the GWAS (OR =4.46, 95% CI: 2.73–7.27, P<0.001) (Table 3) and in the replication study (OR =3.15, 95% CI: 1.40–7.09, P=0.006) (Table 3).
Full table
Full table

Full table
Quality control
Following QC, 459 samples and 21,501 SNPs were identified. The total genotyping rate in the remaining samples was 98.35% (Figure S4A). The PCA plot showed either no stratification or mild stratification of our study population (Figure 1A). The Q-Q plot showed a genomic control inflation factor (λ) of 1.008 (Figure 1B).
Association analysis
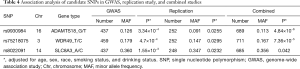
A total of 258 samples met the experimental requirements for the replication study. As shown in Table 4, rs8022091, rs9930984, and rs75218075 were three SNPs found to be significantly associated with the TP53 R249S mutation in the GWAS (MAF =0.36, P=0.00155; MAF =0.126, P=0.000334; MAF =0.179, P=0.00047), in the replication study (MAF =0.347, P=0.0232; MAF =0.091, P=0.0255; MAF =0.147, P=0.0295), and in the combined analysis after adjustment for age, sex, race, smoking status, and drinking status (MAF =0.356, P=0.042; MAF =0.113, P=4.84×10−6; MAF =0.167, P=7.36×10−5). Ultimately, rs8022091, rs9930984, and rs75218075 were identified as SNPs associated with the TP53 R249S mutation and were subjected to further analysis.
Full table
Genetic model analysis
We constructed additive and dominant genetic models to establish the potential associations between genotype variants of the candidate SNPs and the TP53 R249S mutation. These results are shown in Table 5. In the additive genetic model, the TP53 R249S mutation was significantly associated with the TT genotypes of rs9930984 (OR =4.94, 95% CI: 1.48–16.52, P=0.010) and rs75218075 (OR =5.79, 95% CI: 1.33–25.13, P=0.002). In the dominant genetic model, HCC patients carrying the TT genotype of rs9930984 were shown to have a greater risk of TP53 R249S mutation then those carrying GG and GT genotypes (OR =2.27, 95% CI: 1.67–4.44, P=6.43×10−5). A similar result was found in HCC patients carrying the TT genotype of rs75218075, which was associated with a higher risk of TP53 R249S mutation than those carrying CC and CT genotypes (OR =1.98, 95% CI: 1.38–2.85, P=2.16×10−4). However, no significant difference in rs8022091 genotypes was found between the mutation group and non-mutation group in both the additive and dominant genetic models.
Full table
Linkage disequilibrium and haplotype analysis
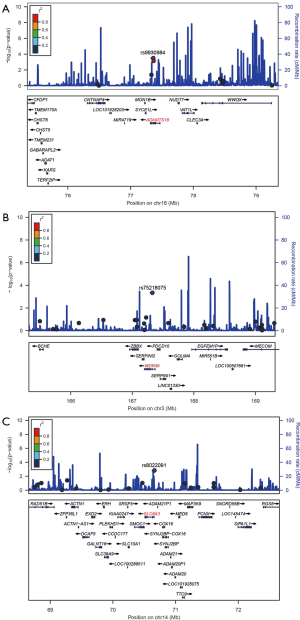
LD and haplotype analysis of SNPs (rs8022091, rs9930984, and rs75218075) was performed within 2 Mb. rs9930984 in the a disintegrin and metallopeptidase with thrombospondin type 1 motif 18 (ADAMTS18) on chromosome 16, within 2 Mb of rs75218075 in WD repeat domain 49 (WDR49) on chromosome 3, and within 2 Mb of rs8022091 in solute carrier family 8 member A3 (SLC8A3) on chromosome 14 (Figure 2A,B,C). Our data shows that rs9930984 is in LD with rs11640912. However, there was no significant association betweenrs11640912 and the TP53 R249S mutation in the replication study (Table S4), and there was no LD found between rs8022091 and rs75218075.
Quantitative RT-PCR
We assessed the mRNA expression of ADAMTS18, WDR49, and SLC8A5 to establish the effects of different SNP genotypes on their gene expression. ADAMTS18 expression was significantly higher in HCC tissue compared with paired non-tumor tissue (P=0.041) (Figure 3A), and populations with the TT genotype of rs9930984 showed significantly less ADAMTS18 expression in non-tumor tissue than individuals with the GT genotype (P=0.0028) (Figure 3A). However, there was no significant difference in ADAMTS18 expression found between GT and TT genotypes of rs9930984 in HCC tissue, or between the TP53 R249S mutation group and the non-mutation group in either HCC tissue or non-tumor tissue (Figure 3A). WDR49 expression was markedly lower in HCC tissue compared with paired non-tumor tissue (P=0.0011). Similarly, we found no significant difference in WDR49 expression between different rs75218075 genotypes, or between the TP53 R249S mutation group and the non-mutation group in either HCC tissue or non-tumor tissue (Figure 3B). No significant difference in SLC8A5 expression was found between HCC tissue or paired non-tumor tissue, between different rs75218075 genotypes, or between the TP53 R249S mutation group and the non-mutation group in non-tumor tissue (Figure 3C).

Discussion
To identify novel loci susceptible to the TP53 R249S mutation in HCC patients both exposed to AFB1 and infected with HBV, we performed a GWAS. A trend of positive TP53 expression was evident in HCC patients with the TP53 R249S mutation compared with HCC patients without this mutation in both the GWAS and the replication study. This was consistent with previous findings (12). Following a combined analysis of all subjects from the GWAS and the subsequent replication study, three SNPs, rs8022091, rs9930984, and rs75218075, remained significantly associated with the TP53 R249S mutation after adjusting for age, sex, race, smoking status, and drinking status.
We further conducted a combined analysis of the genotypes of these three SNPs and the TP53 R249S mutation. Our results suggest that the TT genotype of both rs9930984 and rs75218075 are risk factors associated with the incidence of TP53 R249S mutation in HCC combined with AFB1 exposure and HBV infection. Consistent with these findings, the TT genotypes of rs9930984 and rs75218075 were significantly associated with the TP53 R249S mutation in HCC in the additive genetic model. In contrast, the different genotypes of rs8022091 showed no significant associations with the TP53 R249S mutation in either the additive or dominant model. We performed further analysis for LD within 2 Mb of rs8022091, rs9930984, and rs75218075, and this showed that rs9930984 was in strong LD with rs11640912. However, rs11640912 was not significantly associated with the TP53 R249S mutation in our replication study.
The rs9930984 SNP is located in the ADAMTS18 gene, which encodes a member of the ADAMTS protein family. ADAMTS18 has been identified as a novel functional tumor suppressor (29). More and more studies (29-33) have found that mutations in this gene, as well as hypermethylation of promoters, is closely related to the growth of multiple types of tumors. This suggests that ADAMTS18 plays a role in tumor suppression. Compared with paired adjacent non-tumor tissue or normal tissue, ADAMTS18 CpG methylation is significantly upregulated in gastric, colorectal, and pancreatic cancer tissue (31). Pharmacological and genetic demethylation analyses indicate that CpG methylation directly inhibits the expression of ADAMTS18. In vitro studies have found that the restoration of ADAMTS18 gene expression previously downregulated or silenced in tumor cell lines inhibits tumor cell clonality (29). However, a study on malignant melanoma showed that mutated ADAMTS18 promoted cell growth, migration, and metastasis (32). In our study, ADAMTS18 rs9930984 was significantly correlated with the TP53 R249S mutation, and participants carrying the TT genotype of rs9930984 showed a higher risk of TP53 R249S mutation compared with those carrying GG and GT genotypes. The results of our separate experiments both showed significantly higher ADAMTS18 mRNA expression in HCC tissue compared with non-tumor tissue. However, no differences in ADAMTS18 expression were found between the TP53 R249S mutation group and the non-mutation group for both tumor tissue and non-tumor tissue. ADAMTS18 expression in non-tumor tissue with the rs9930984 TT genotype was lower than that expressed in tissue with the GT genotype. To our knowledge, no studies to date have demonstrated an association between ADAMTS18 and HCC pathogenesis or the TP53 R249S mutation. Moreover, no relevant research on rs9930984 and its potential role in HCC is presently documented.
The rs75218075 SNP is located in the WDR49 gene on chromosome 3, which encodes a member of the WDR protein family with nine WD repeats. These proteins are involved in multiple cellular processes, including cell cycle progression, signal transduction, apoptosis, and gene regulation (34,35). WDR79 is reportedly involved in the development and progression of non-small cell lung cancer (36). A previous in vitro study on bladder cancer by Chen et al. (37) showed that WDR5 is upregulated in tumor tissue and promotes the proliferation of bladder cancer cells and the autologous renewal of cells, as well as increasing chemoresistance. However, no studies on the biological function of WDR49 have been reported to date, and its role in tumor development remains unknown. Our results suggest that WDR49 rs75218075 is significantly associated with the TP53 R249S mutation (Table 4, Tables S2,S4), and patients carrying the rs75218075 TT genotype might be at greater risk of expressing the TP53 R249S mutation compared with those carrying CC and CT genotypes. WDR49 mRNA expression was significantly lower in HCC tissue compared with non-tumor tissue, but was not correlated with any genotypes of rs75218075. We also found no correlation between WDR49 mRNA expression and either the TP53 R249S mutation group or non-mutation group in either HCC or non-tumor tissue.
The rs8022091 SNP is located in the SLC8A gene on chromosome 14 and encodes a member of the sodium/calcium exchanger integral membrane protein family. Recent studies have shown that SLC8A3 is an essential component of PKCα activation by Ca2+ in endothelial cell membranes, a necessary step of VEGF-induced ERK1/2 phosphorylation and angiogenesis (38,39). Reverse transport of SLC8A3 has been reported in cardiac fibroblasts (39) and neuroblastoma cells (40); reverse transport is required for ERK1/2 activation. A recent study showed a significant negative correlation between promoter CpG island methylation and SLC8A3 gene expression in clear-cell renal cell cancer (41), indicating that the SLC8A3 gene may play a functional epigenetic control role in tumors. In both our GWAS and replication study, SLC8A3 rs8022091 was significantly associated with the TP53 R249S mutation, though there was no correlation between any rs8022091 genotypes and the TP53 R249S mutation.
In addition, TP53 mutations are reportedly associated with the prognosis of multiple cancers and are biomarkers of environmental carcinogens. Our previous study has shown that patients with TP53 R249S mutation have a higher risk of recurrence within 2 years after surgery in HBV-relative HCC (42). Nogueira et al. showed that TP53 R249S mutation was associated with tumor diameter and tumor differentiation in Brazilian HCC patients (43). However, in our previous study, we did not find such those association, possibly because our study population was all HBV-associated HCC, and their study population included people with HBV, HCV and non-hepatitis virus backgrounds. Moreover, their study sample was only 53, and the reliability of their results was questionable. Several studies on Japanese HCC patients have shown (35,36) that TP53 gene mutation is significantly correlated with short-term tumor recurrence and tumor differentiation, which was similar to our previous results (44,45). Other studies in China have also shown that TP53 R249S mutation is an indicator of poor prognosis in HCC (46,47). Hence, TP53 R249S mutations may be an independent prognostic biomarker for HCC.
In our study, no significant differences were detected between SLC8A3 mRNA expression and either HCC tissue or non-tumor tissue. We also found no associations between SLC8A3 mRNA expression and any rs8022091 genotypes, or SLC8A3 mRNA expression and either the TP53 R249S mutation group or non-mutation group in either HCC tissue or non-tumor tissue.
Conclusions
We successfully conducted a comprehensive GWAS and replication study on HCC patients exposed to AFB1 and HBV in the Guangxi region. The SNPs ADAMTS18 rs9930984 and WDR49 rs75218075 were found to be associated with susceptibility to the TP53 R249S mutation, and the TT genotypes of rs9930984 and rs75218075 were further identified as risk factors for this mutation. Further large-scale, multi-center studies are necessary to verify our findings and establish the specific associations between these regions and this codon 249 TP53 gene mutation in HCC patients exposed to both AFB1 and HBV.
Acknowledgments
The authors thank Prof. Ya Guo, Zhixiong Su, Ming Su, Bin Chen, Zhang Wen, and Banghao Xu for contributing to this study through the collection of samples.
Funding: This work was supported by the National Nature Science Foundation of China (Grant Nos. 81802874, 81560535, and 81072321), the Natural Science Foundation of the Guangxi Province of China (Grant No. 2018 GXNSFBA138013), the Guangxi Key R&D Program (GKEAB18221019), the 2009 Program for New Century Excellent Talents in University (NCET), Natural Science Foundation of Guangxi Province of China (Grant No. 2020GXNSFAA159127), the Guangxi Nature Sciences Foundation (No.: GuiKeGong 1104003A-7), the Guangxi Health Ministry Medicine Grant (Key-Scientific Research-Grant Z201018), the Self-raised Scientific Research Fund of the Guangxi Zhuang Autonomous Region Health and Family Planning Commission (Z2016318), the Key Laboratory of High-Incidence-Tumor Prevention & Treatment (Guangxi Medical University), and the Ministry of Education (GKE2018-01).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/jgo-20-510
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jgo-20-510
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jgo-20-510). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. These studies were approved by the Ethical Review Committee of the First Affiliated Hospital of Guangxi Medical University {Approval Number: 2015 [KY-E-072]}. All participants signed written informed consent. This study conformed to the provisions of the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Mittal S, El-Serag HB. Epidemiology of hepatocellular carcinoma: consider the population. J Clin Gastroenterol 2013;47 Suppl:S2-S6. [Crossref] [PubMed]
- Pocha C, Xie C. Hepatocellular carcinoma in alcoholic and non-alcoholic fatty liver disease—one of a kind or two different enemies? Transl Gastroenterol Hepatol 2019;4:72. [Crossref] [PubMed]
- Wei B, Chen JS, Yang SL, et al. A scoring model combining serum alpha-fetoprotein and tumor size and number predicts prognosis in hepatitis B virus-related hepatocellular carcinoma patients after curative hepatectomy. Transl Cancer Res 2019;8:1438-48. [Crossref]
- Huy TTT, Abe K. Molecular epidemiology of hepatitis B and C virus infections in Asia. Pediatr Int 2004;46:223-30. [Crossref] [PubMed]
- Kujawa M. Some Naturally Occurring Substances: Food Items and Constituents, Heterocyclic Aromatic Amines and Mycotoxins. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, Vol. 56. Herausgegeben von der International Agency for Research on Cancer, Wo. Mol Nutr Food Res 2010;38:351.
- Fan JH, Wang JB, Jiang Y, et al. Attributable causes of liver cancer mortality and incidence in china. Asian Pac J Cancer Prev 2013;14:7251-6. [Crossref] [PubMed]
- Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013;502:333-9. [Crossref] [PubMed]
- Robles AI, Harris CC. Clinical outcomes and correlates of TP53 mutations and cancer. Cold Spring Harb Perspect Biol 2010;2:a001016. [Crossref] [PubMed]
- Liu ZM, Li LQ, Peng MH, et al. Hepatitis B virus infection contributes to oxidative stress in a population exposed to aflatoxin B1 and high-risk for hepatocellular carcinoma. Cancer Lett 2008;263:212-22. [Crossref] [PubMed]
- Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet 2012;379:1245-55. [Crossref] [PubMed]
- Qi LN, Bai T, Chen ZS, et al. The p53 mutation spectrum in hepatocellular carcinoma from Guangxi, China: role of chronic hepatitis B virus infection and aflatoxin B1 exposure. Liver Int 2015;35:999-1009. [Crossref] [PubMed]
- Smela ME, Currier SS, Bailey EA, et al. The chemistry and biology of aflatoxin B(1): from mutational spectrometry to carcinogenesis. Carcinogenesis 2001;22:535-45. [Crossref] [PubMed]
- Woo HG, Wang XW, Budhu A, et al. Association of TP53 mutations with stem cell-like gene expression and survival of patients with hepatocellular carcinoma. Gastroenterology 2011;140:1063-70. [Crossref] [PubMed]
- Kew MC. Synergistic interaction between aflatoxin B1 and hepatitis B virus in hepatocarcinogenesis. Liver Int 2003;23:405-9. [Crossref] [PubMed]
- Liu Z, Hu Y, Yang X, et al. Combinative analysis of factors influence serum alanine aminotransferase activity in adult male population from southern China. Clin Biochem 2012;45:1683-8. [Crossref] [PubMed]
- Kew MC. Hepatitis B virus x protein in the pathogenesis of hepatitis B virus-induced hepatocellular carcinoma. J Gastroenterol Hepatol 2011;26 Suppl 1:144-52. [Crossref] [PubMed]
- Jiang W, Wang XW, Unger T, et al. Cooperation of tumor-derived HBx mutants and p53-249(ser) mutant in regulating cell proliferation, anchorage-independent growth and aneuploidy in a telomerase-immortalized normal human hepatocyte-derived cell line. Int J Cancer 2010;127:1011-20. [Crossref] [PubMed]
- Yeh FS, Yu MC, Mo CC, et al. Hepatitis B virus, aflatoxins, and hepatocellular carcinoma in southern Guangxi, China. Cancer Res 1989;49:2506-9. [PubMed]
- Visscher PM, Brown MA, McCarthy MI, et al. Five years of GWAS discovery. Am J Hum Genet 2012;90:7-24. [Crossref] [PubMed]
- Bruix J, Sherman M. Management of hepatocellular carcinoma. Hepatology (Baltimore, Md) 2005;42:1208-36. [Crossref] [PubMed]
- Pugh RN, Murray-Lyon IM, Dawson JL, et al. Transection of the oesophagus for bleeding oesophageal varices. Br J Surg 1973;60:646-9. [Crossref] [PubMed]
- Kondo K, Chijiiwa K, Kai M, et al. Surgical strategy for hepatocellular carcinoma patients with portal vein tumor thrombus based on prognostic factors. J Gastrointest Surg 2009;13:1078-83. [Crossref] [PubMed]
- Yu L, Liu X, Han C, et al. XRCC1 rs25487 genetic variant and TP53 mutation at codon 249 predict clinical outcomes of hepatitis B virus-related hepatocellular carcinoma after hepatectomy: A cohort study for 10 years' follow up. Hepatol Res 2016;46:765-74. [Crossref] [PubMed]
- Cote RJ, Dunn MD, Chatterjee SJ, et al. Elevated and absent pRb expression is associated with bladder cancer progression and has cooperative effects with p53. Cancer Res 1998;58:1090-4. [PubMed]
- Esrig D, Elmajian D, Groshen S, et al. Accumulation of nuclear p53 and tumor progression in bladder cancer. N Engl J Med 1994;331:1259-64. [Crossref] [PubMed]
- Lin DY, Tang ZZ. A general framework for detecting disease associations with rare variants in sequencing studies. Am J Hum Genet 2011;89:354-67. [Crossref] [PubMed]
- Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 2010;26:2336-7. [Crossref] [PubMed]
- Jin H, Wang X, Ying J, et al. Epigenetic identification of ADAMTS18 as a novel 16q23.1 tumor suppressor frequently silenced in esophageal, nasopharyngeal and multiple other carcinomas. Oncogene 2007;26:7490-8. [Crossref] [PubMed]
- Nordgard SH, Johansen FE, Alnaes GI, et al. Genome-wide analysis identifies 16q deletion associated with survival, molecular subtypes, mRNA expression, and germline haplotypes in breast cancer patients. Genes Chromosomes Cancer 2008;47:680-96. [Crossref] [PubMed]
- Li Z, Zhang W, Shao Y, et al. High-resolution melting analysis of ADAMTS18 methylation levels in gastric, colorectal and pancreatic cancers. Med Oncol 2010;27:998-1004. [Crossref] [PubMed]
- Wei X, Prickett TD, Viloria CG, et al. Mutational and functional analysis reveals ADAMTS18 metalloproteinase as a novel driver in melanoma. Mol Cancer Res 2010;8:1513-25. [Crossref] [PubMed]
- Wagstaff L, Kelwick R, Decock J, et al. The roles of ADAMTS metalloproteinases in tumorigenesis and metastasis. Front Biosci (Landmark Ed) 2011;16:1861-72. [Crossref] [PubMed]
- Neer EJ, Schmidt CJ, Nambudripad R, et al. The ancient regulatory-protein family of WD-repeat proteins. Nature 1994;371:297-300. [Crossref] [PubMed]
- Smith TF, Gaitatzes C, Saxena K, et al. The WD repeat: a common architecture for diverse functions. Trends Biochem Sci 1999;24:181-5. [Crossref] [PubMed]
- Sun Y, Yang C, Chen J, et al. Overexpression of WDR79 in non-small cell lung cancer is linked to tumour progression. J Cell Mol Med 2016;20:698-709. [Crossref] [PubMed]
- Chen X, Xie W, Gu P, et al. Upregulated WDR5 promotes proliferation, self-renewal and chemoresistance in bladder cancer via mediating H3K4 trimethylation. Sci Rep 2015;5:8293. [Crossref] [PubMed]
- Andrikopoulos P, Fraser SP, Patterson L, et al. Angiogenic functions of voltage-gated Na+ Channels in human endothelial cells: modulation of vascular endothelial growth factor (VEGF) signaling. J Biol Chem 2011;286:16846-60. [Crossref] [PubMed]
- Andrikopoulos P, Baba A, Matsuda T, et al. Ca2+ influx through reverse mode Na+/Ca2+ exchange is critical for vascular endothelial growth factor-mediated extracellular signal-regulated kinase (ERK) 1/2 activation and angiogenic functions of human endothelial cells. J Biol Chem 2011;286:37919-31. [Crossref] [PubMed]
- Nashida T, Takuma K, Fukuda S, et al. The specific Na(+)/Ca(2+) exchange inhibitor SEA0400 prevents nitric oxide-induced cytotoxicity in SH-SY5Y cells. Neurochem Int 2011;59:51-8. [Crossref] [PubMed]
- Deckers IA, van Engeland M, van den Brandt PA, et al. Promoter CpG island methylation in ion transport mechanisms and associated dietary intakes jointly influence the risk of clear-cell renal cell cancer. Int J Epidemiol 2017;46:622-31. [PubMed]
- Qin W, Han C, Mai R, et al. Establishment of a prognostic model for predicting short-term disease-free survival in cases of hepatitis B-related hepatocellular carcinoma with the TP53 249Ser mutation in southern China. Transl Cancer Re 2020;9:4517-33. [Crossref]
- Nogueira JA, Ono-Nita SK, Nita ME, et al. 249 TP53 mutation has high prevalence and is correlated with larger and poorly differentiated HCC in Brazilian patients. BMC Cancer 2009;9:204. [Crossref] [PubMed]
- Hayashi H, Sugio K, Matsumata T, et al. The clinical significance of p53 gene mutation in hepatocellular carcinomas from Japan. Hepatology 1995;22:1702-7. [Crossref] [PubMed]
- Sugo H, Takamori S, Kojima K, et al. The significance of p53 mutations as an indicator of the biological behavior of recurrent hepatocellular carcinomas. Surg Today 1999;29:849-55. [Crossref] [PubMed]
- Su H, Zhao J, Xiong Y, et al. Large-scale analysis of the genetic and epigenetic alterations in hepatocellular carcinoma from Southeast China. Mutat Res 2008;641:27-35. [Crossref] [PubMed]
- Yuan RH, Chang KT, Chen YL, et al. S100P expression is a novel prognostic factor in hepatocellular carcinoma and predicts survival in patients with high tumor stage or early recurrent tumors. PLoS One 2013;8:e65501. [Crossref] [PubMed]
(English Language Editor: B. Madden)


