
Genomic characteristics in neoadjuvant chemoradiotherapy for locally advanced esophageal squamous cell carcinoma
Introduction
Esophageal cancer ranks as the sixth largest contributor to cancer mortality in the world, with East Asia having the highest prevalence of the disease (1). Among Chinese esophageal cancer patients, esophageal squamous cell carcinoma (ESCC) is the dominant pathological type, comprising >90% of all cases. Most ESCC patients have already progressed to a locally advanced stage when they are first diagnosed. Therefore, it remains a great challenge to achieve a favorable prognosis through surgery alone, and the 5-year survival rate stands at 20–30% (2).
Increasing clinical evidence suggests that neoadjuvant chemoradiotherapy (nCRT) followed by esophagectomy can significantly increase both the overall and disease-free survival of ESCC patients (3). It has also been reported that 43–49% of ESCC patients could obtain a pathological complete response (pCR), with a lower rate of recurrence (4,5). However, almost 30% of ESCC patients have been found to have little or no response to nCRT and thus might be at risk of disease progression, delayed resection, and unnecessary exposure to nCRT toxicity. Despite NCRT lacking efficacy for a considerable proportion of ESCC patients, an effective biomarker for choosing the patient population that could benefit from chemoradiotherapy has yet to be discovered. In addition, genes play a critical role in the oncogenesis, progression, and resistance of ESCC. Therefore, it is extremely important to identify biomarkers to predict the outcome of nCRT and to investigate the mechanism of resistance therapy.
In this study, we investigated genomic differences between patients with locally advanced ESCC who achieved pathologic complete response (pCR) and those who showed little or no response (pSD) to nCRT by analyzing whole-exome sequencing (WES) data from 27 paired samples of ESCC tissue before and following nCRT. This article is presented in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/jgo-20-504).
Methods
DNA extraction and quantification
We retrospectively collected pretreatment gastroscopic biopsy tissue and postoperative tissue obtained from patients with ESCC to create formalin-fixed, paraffin-embedded (FFPE) tissue samples. The FFPE blocks were twice subjected to xylene deparaffinization, after which the QIAamp DNA FFPE Tissue Kit (Qiagen) was employed for the extraction of DNA, in line with the protocol supplied by the manufacturer. Xylene is used to remove FFPE sections and cores, which were then washed with ethanol. Proteinase K was used to digest the tissues at 56 °C overnight, followed by incubation at 90 °C for 5 minutes for DNA crosslink reversal . Genomic DNA was isolated from tissue samples using Nanodrop2000 (Thermo), and DNA quantification was performed using Qubit3.0 (Life Technology) with a dsDNA HS Assay Kit (Life Technology) according to the manufacturers’ instructions. Approval for this study was granted by the Sichuan Cancer Hospital Ethical Committee (NO. SCCHEC-02-2017-043), and informed consent forms were signed by every patient. The study conformed to the provisions of the Declaration of Helsinki (as revised in 2013).
Library preparation and sequencing
DNA libraries were amplified through polymerase chain reaction (PCR) and purified prior to targeted enrichment. Libraries from different samples were marked with unique indices, and up to 2 µg of different libraries was pooled together for targeted enrichment. Nonspecific binding of library DNA to targeted probes was blocked through the addition of human Cot-1 DNA (Life Technologies) and xGen Universal Blocking Oligos (Integrated DNA Technologies). WES was performed on an Illumina HiSeq 2000 system. Paired-end sequencing with a read length of 100 base pairs and 90× average on-target coverage was conducted.
Bioinformatics analysis
Original image data were acquired using the Illumina HiSeq 2000 sequencing platform, and base calling analysis was performed to convert them to raw sequence data, which contained sequence information and corresponding sequencing quality information. Identification of single-nucleotide variants (SNVs) and short insertions/deletions (indels) was performed using VarScan2 with the minimum variant allele frequency threshold set at 0.01, and P value threshold for calling variants set at 0.05 for the generation of files in variant call format . The annotation of each SNV/indel was carried out with ANNOVAR, and the Integrative Genomics Viewer was employed for manual checking of the SNVs/indels. ANNOVAR was also used to annotate single-nucleotide polymorphisms (SNPs) and indels against the dbSNP (v138), 1000Genome, and ESP6500 (population frequency >0.015) databases. Only missense, stopgain, frameshift and non-frameshift indel mutations were kept. Copy number variation (CNV) detection was performed with custom-made software.
Statistical analysis
For biomarker analysis, the significance with categorical variables were assessed by Fisher exact test. Continuous variables that conformed to the normal distribution were implemented by student’s t-test. If the data failed to meet the normal distribution, non-parametric analyses, Mann-Whitney U test and Wilcoxon’s rank-sum test were used. All above tests were two-sided and the statistically significant was set to a P value of less than 0.05. The statistical analysis was conducted using IBM SPSS Statistics 22 or R (version).
Results
Patient characteristics
All patients in this study had been diagnosed as locally potentially resectable ESCC clinically staged as T3-4aN0-2M0 according to the 8th edition of the American Cancer Commission (AJCC) TNM staging system (6) before treatment. Those assessed as being suitable for nCRT were given intravenous (IV) administration of carboplatin 300 mg/m2 on day 1 and paclitaxel 135 mg/m2 IV administration on day 1, every 3 weeks for 2 cycles, as well as concurrent radiation with a total dose of 40.0 Gy delivered in 20 fractions of 2.0 Gy 5 days per week. Individuals without progression to unresectable or metastatic disease underwent esophagectomy 4–6 weeks later. Fourteen patients, including 7 cases of pCR and 7 of pSD, were enrolled in this study. There was no selection bias in the clinical characteristics of the patients in each group, such as age, sex, tumor location, and cellular differentiation (Table 1). During a median follow-up of 26 months, 1 pCR and 4 pSD patients died. A total of 28 FFPE samples were used for WES, with an average sequencing depth of 90× (range, 31–137×). One of the pretreatment samples (Case B) failed quality control. The WES data of the remaining 27 samples were analyzed.
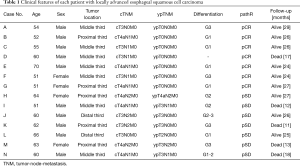
Full table
Mutation landscape in ESCC
The WES revealed 148 pan-cancer driver genes in all 27 samples (Figure 1). TP53 was the most frequently detected gene, accounting for 44% of ESCC (12/27). Other frequently mutated ESCC driver genes included KMT2D (10/27, 37%), NOTCH2 (9/27, 33%), KMT2A (6/27, 22%), PIK3CA (6/27, 22%), ATR (5/27, 19%), FAT1 (5/27, 19%), HUWE1 (5/27, 19%), PTPRD (5/27, 19%), EPHA3 (4/27, 15%), FBXW7 (4/27, 15%), NCOR1 (4/27, 15%), NF1 (4/27, 15%), and NIPBL (4/27, 15%). Among them, TP53, KMT2D, NOTCH2, KMT2A, PIK3CA, FAT1, FBXW7, and NOTCH1 were the common driver genes in ESCC, which was consistent with results of previous studies (7-9).
CNVs
Somatic CNVs were detected in 22 samples (Figure 2). Overall, copy number loss was detected more frequently than copy number gain. In the pretreatment samples, loss of chromosome 19p13.11 was detected more often in the pretherapy samples of the pCR group (5/6) than in those of the non-responders (1/7) (P=0.0291, Fisher’s exact test). Meanwhile, a significant increase in the frequency of 19q13.31 gain was observed among the post-therapy samples (5/14). The CNV of these chromosome fragments might have affected the therapeutic efficacy.
Diver gene mutation
KMT2A missense mutation was only detected in 3 of 6 pretreatment samples in the pSD group, and it did not appear in the pretreatment samples of the pCR group (P=0.07, Fisher’s exact test). Frame shift and nonsense mutations in KMT2A were found in only the postoperative samples in each group. We also found a new NF1 variation in 4 ESCC patients (1 in the pCR group, 3 in the pSD group) after nCRT, which had not been detected in their pretreatment samples. Other mutant genes such as KMT2D, NOTCH2, and NIPBL appeared more frequently in post-therapy samples.
Next, we analyzed the sequencing data to identify differences in the mutated genes between before and after therapy (Table 2). Although a few mutant genes (from 4 to 25) were found to be coincident, of more than 185 genes (up to 758 genes) in the pCR group (A-G), no more than 2 genes had the same sites. Similarly, 5 paired samples from the pSD group, had only 1 or even no overlapping genes. Based on this result, the gene landscape was totally different between the pre- and post-treatment samples, which suggested that tumor genes had great heterogeneity among the ESCC patients after nCRT.
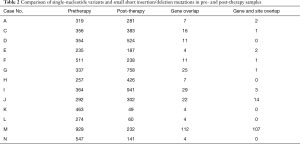
Full table
The SNV spectra of all patients’ samples are shown in Figure 3. They revealed statistically significant reductions in C/G>G/C (P=0.003) and C/G>A/T (P=0.002) transitions from before to after treatment. The same results were independently obtained for the pCR group (C/G>G/C, P=0.027; C/G>A/T, P=0.004) and the pSD group (C/G>G/C, P=0.032; C/G>A/T, P=0.017).
Discussion
We analyzed paired tissue samples obtained before and after nCRT and found that chromosome 19p13.11 loss in the pretreatment tissue samples could be a positive predictor of pCR after nCRT. Patients with a KMT2A missense mutation before treatment tended not to benefit from nCRT. This is 1 of the first reports to compare the genomic differences between patients with ESCC who achieve pCR and those with pSD after nCRT.
Several recent publications have assessed the pathologic markers of responders to nCRT. A prospective study using diffusion-weighted magnetic resonance imaging showed that a higher post-nCRT apparent diffusion coefficient value could be a predictive indicator of good pathologic response to nCRT in ESCC patients (10). Another retrospective exploratory study suggested that higher PD-L1 expression and the CD8+ and CD4+ lymphocyte ratio were more relevant to pCR in a cohort of 88 ESCC patients receiving nCRT (11), whereas another study disclosed the opposite result that positive PD-L1 expression predicted a poor nCRT response (12). Recently, we reported the results of an analysis of the NEOCRTEC5010 trial, which revealed that persistent pathologic lymph node metastasis after NCRT was significantly associated with a poor prognosis in ESCC patients (13).
All of these studies systematically explored predictive factors for different pathologic responses to nCRT in ESCC patients through image inspection, clinical and pathological features, and immunohistochemical profiles. Our study revealed for the first time that chromosome CNV may be a predictor of the pathological response to nCRT for ESCC.
Previous studies have suggested that CNVs on 19p13.11 are related to patient’s characteristics and prognosis of cancer (14,15). Copy number loss on 19p13.11 is more often observed in lung adenocarcinoma in Caucasian patients (14). Patients with ovarian cancer who harbored copy number gain on 19p13.11 were reported to have poor overall and progression-free survival (15). To the best of our knowledge, no previous study has implicated 19p13.11 loss with the efficacy of chemoradiotherapy.
One of limitations in this study is the small number of recruited patient samples; our results need to be verified in an analysis involving a larger number of samples. In addition, bias was introduced with pretreatment samples obtained by puncture, which may have affected the results. We also did not investigate gene expression, so we might have missed a lot of valuable information.
In conclusion, we observed that 19p13.11 copy number loss and KMT2A missense mutation might be biomarkers to predict the outcome of pCR; however, further validation in a prospective study of a larger sample is required.
Acknowledgments
The authors thank Lihua Chen for his direct on the experimental design, and Depei Huang, Xudong Shen, Guofeng Zhao, and Jianqing Yang for their assistance in statistical analysis and interpretation.
Funding: This work was supported by Sichuan Science and Technology Program (Grant No. 2018SZ0199, 2019JDRC0076).
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/jgo-20-504
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jgo-20-504
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jgo-20-504). All authors report grants from Sichuan Science and Technology Program, during the conduct of the study.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Approval for this study was granted by the Sichuan Cancer Hospital Ethical Committee (No. SCCHEC-02-2017-043), and informed consent forms were signed by every patient. The study conformed to the provisions of the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Ma WJ, Zhang QN, Shi SZ, et al. Preoperative chemoradiation may be more effective for esophageal squamous cell carcinoma compared with adenocarcinoma: results from 15 randomized controlled trials of 2,250 patients. Transl Cancer Res 2018;7:1421-10. [Crossref]
- van Hagen P, Hulshof MC, van Lanschot JJ, et al. Preoperative chemoradiotherapy for esophageal or junctional cancer. N Engl J Med 2012;366:2074-84. [Crossref] [PubMed]
- Shapiro J, van Lanschot JJB, Hulshof M, et al. Neoadjuvant chemoradiotherapy plus surgery versus surgery alone for oesophageal or junctional cancer (CROSS): long-term results of a randomised controlled trial. Lancet Oncol 2015;16:1090-8. [Crossref] [PubMed]
- Yang H, Liu H, Chen Y, et al. Neoadjuvant Chemoradiotherapy Followed by Surgery Versus Surgery Alone for Locally Advanced Squamous Cell Carcinoma of the Esophagus (NEOCRTEC5010): A Phase III Multicenter, Randomized, Open-Label Clinical Trial. J Clin Oncol 2018;36:2796-803. [Crossref] [PubMed]
- Rice TW, Ishwaran H, Ferguson MK, et al. Cancer of the Esophagus and Esophagogastric Junction: An Eighth Edition Staging Primer. J Thorac Oncol 2017;12:36-42.
- Gao YB, Chen ZL, Li JG, et al. Genetic landscape of esophageal squamous cell carcinoma. Nat Genet 2014;46:1097-102. [Crossref] [PubMed]
- Sawada G, Niida A, Uchi R, et al. Genomic Landscape of Esophageal Squamous Cell Carcinoma in a Japanese Population. Gastroenterology 2016;150:1171-82. [Crossref] [PubMed]
- Hu N, Kadota M, Liu H, et al. Genomic Landscape of Somatic Alterations in Esophageal Squamous Cell Carcinoma and Gastric Cancer. Cancer Res 2016;76:1714-23. [Crossref] [PubMed]
- Li QW, Qiu B, Wang B, et al. Prediction of pathologic responders to neoadjuvant chemoradiotherapy by diffusion-weighted magnetic resonance imaging in locally advanced esophageal squamous cell carcinoma: a prospective study. Dis Esophagus 2018. [Crossref] [PubMed]
- Fassan M, Cavallin F, Guzzardo V, et al. PD-L1 expression, CD8+ and CD4+ lymphocyte rate are predictive of pathological complete response after neoadjuvant chemoradiotherapy for squamous cell cancer of the thoracic esophagus. Cancer Med 2019;8:6036-48. [Crossref] [PubMed]
- Huang WT, Lu HI, Wang YM, et al. Positive Programmed Cell Death-Ligand 1 Expression Predicts Poor Treatment Outcomes in Esophageal Squamous Cell Carcinoma Patients Receiving Neoadjuvant Chemoradiotherapy. J Clin Med 2019;8:1864. [Crossref] [PubMed]
- Leng X, He W, Yang H, et al. Prognostic Impact of Postoperative Lymph Node Metastases After Neoadjuvant Chemoradiotherapy for Locally Advanced Squamous Cell Carcinoma of Esophagus: From the Results of NEOCRTEC5010, a Randomized Multicenter Study. Ann Surg 2019. Epub ahead of print. [Crossref] [PubMed]
- Broet P, Dalmasso C, Tan EH, et al. Genomic profiles specific to patient ethnicity in lung adenocarcinoma. Clin Cancer Res 2011;17:3542-50. [Crossref] [PubMed]
- Sung CO, Song IH, Sohn I. A distinctive ovarian cancer molecular subgroup characterized by poor prognosis and somatic focal copy number amplifications at chromosome 19. Gynecol Oncol 2014;132:343-50. [Crossref] [PubMed]
(English Language Editor: K. Brown)





