
N-myc downstream-regulated gene 2 promotes the protein stability of estrogen receptor beta via inhibition of ubiquitin-protein ligase E3A to suppress colorectal cancer
Introduction
Colorectal cancer (CRC) is the third most common malignancy and the second leading cause of cancer-related death worldwide (1). Despite the slow progression of CRC and good clinical outcomes associated with early-stage diagnosis, preventive methods and effective treatment are currently inadequate (2). Overall, female patients with CRC have lower odd ratios and longer overall survival (OS) than males; however, older women are more susceptible to CRC and have a lower OS than men (3). The sex differences in outcomes of CRC patients might be attributable to various activated states of estrogen and its receptor-related signaling pathways.
Estrogen is a steroid hormone that has multiple functions, which include binding to estrogen receptors (ERs) to regulate downstream genes. There is high sequence and similar structural homology between the transcription factors ERα and ERβ (encoded by ESR1/ESR2 gene), both of which are activated by ligands and in turn activate downstream genes to elicit a biological response (4). The overexpression of ERα can stimulate the occurrence of precancerous lesions and breast cancer (5), while a high expression of ERβ inhibits cellular proliferation and promotes CRC cell apoptosis (6,7). An increasing number of reports have shown that in CRC patients, a lower expression of ERβ is associated with a more advanced stage (8,9). Taken together, these reports suggest that ERβ might be a good prognostic indicator in CRC. However, the exact molecular mechanism of ERβ in tumor suppressor genes is unclear. Thus, investigation of the key molecules in the regulatory network of ERβ in CRC patients is vital.
N-myc downstream-regulated gene (NDRG)-2 is a member of the NDRG protein family, which is involved in both cellular differentiation and tumor suppression in the intestinal epithelium (10). Compared with that in normal tissues, NDRG2 expression is decreased in colorectal, gastric (11), breast and liver (12) cancer tissues. Moreover, in CRC patients, the expression level of NDRG2 messenger RNA (mRNA) is closely associated with tumor differentiation status, lymph node metastasis (13), and prognosis (14). Consequently, NDRG2 expression might have potential as a therapeutic target in CRC. Intriguingly, a novel report found that estradiol (E2) or diarylpropionitrile (DPN) could up-regulate NDRG2 at both the mRNA and protein levels in a dose- and time-dependent manner (15). In addition, we found that NDRG2 could improve ERβ protein stability in CRC cells. However, there is still no direct evidence to verify the relationship between NDRG2 and ERβ. Specifically, the molecular mechanism and biological function of the regulatory loop between NDRG2 and ERβ demand further investigation.
The aim of the study is to explore the role of NDRG2 in ERβ and their effects on progression of CRC. We demonstrate that NDRG2 is clinically associated with ERβ and NDRG2 up-regulates the protein stability of ERβ without affecting its mRNA levels. Mechanistically, NDRG2 down-regulates ubiquitination degradation of ERβ by inhibiting the ability of UBE3A to combine with ERβ. NDRG2high/ESR2high is a signature correlated with good outcome in CRC patients and could be used as a prognostic marker in CRC. We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/jgo-20-557).
Methods
Patient samples and tissue microarray analysis
Tissue samples from 34 CRC patients from Xijing Digestive Hospital were used in this study. Tissue microarrays were provided by the Department of Pathology in The Fourth Military Medical University. Thirty-four pairs of tumor tissue samples and corresponding adjacent normal mucosa samples were included in the tissue microarray. An immunohistochemical (IHC) assay was conducted using specific antibodies of NDRG2 (CST; 5667S; 1:1,000) and ERβ (Abcam; ab3576; 1:1,000). IHC images were scanned by Panoramic MIDI (Santa Clara, CA, USA), and adjusted with a histochemical score (H-Score) by Quant Center. Correlation analysis of NDRG2 and ERβ was using GraphPad Prism (Version 6; La Jolla, CA, USA).
All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Xijing Hospital Ethics Committee (No. KY20203269-1) and informed consent was taken from all the patients.
Clinical data analysis
The gene expression profiles and survival data of CRC patients were retrieved from The Cancer Genome Atlas (TCGA) clinical database (https://portal.gdc.cancer.gov/). The TCGA dataset comprised 39 paracancerous tissue samples and 398 cancerous tissue samples, and complete follow-up data were available for 379 patients.
Gene set enrichment analysis
To explore the potential biological functions of NDRG2 and ESR2, gene set enrichment analysis (GSEA) was performed with GSEA version 4.0.1 software. According to the expression of NDRG2/ESR2, all CRC samples were divided into two groups based on the median value of genes. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway gene set was used as the reference gene set.
Cell lines and reagents
The human colon cancer cell lines HCT116 and HT29 were purchased from the American Type Culture Collection (Manassas, VA, USA) and cultured in McCoy’s 5A medium (BOSTER) with 10% of fetal calf serum and 1% penicillin/streptomycin. SW620 cells were cultured in HyClone Leibovitz L-15 medium (BOSTER), and HEK293T cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Gibco-BRL). Control or NDRG2-overexpression cells were constructed in the same vectors (pcDNA3.1, pcDNA3.1-NDRG2) as we reported (16). DPN and the proteasome inhibitor MG132 were purchased from MedChemExpress (MCE). CHX (cycloheximide) was purchased from Sigma-Aldrich. CRC cells were treated with 0–1,000 nmol/L DPN for 24–96 hours before being harvested for downstream assays. The doses of CHX and MG132 used to treat the CRC cell lines were 20 µg/mL and 50 nmol/mL, respectively.
Western blotting
For detection, the protein levels of NDRG2, ERβ, UBE3A, Tubulin, LaminA/C, MYC-tag, Flag tag, V5-ub and β-actin, cells were collected after washing by phosphate-buffered saline (PBS) three times, and then disrupted by RIPA Lysis Buffer. Equal amounts of protein were quantified using a BCA™ protein assay kit. Proteins with different molecular weights were separated by sodium odecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE), and then transferred to nitrocellulose membranes. After blocking with 5% nonfat milk, the membranes were incubated with antibodies against NDRG2 (CST; 5667S; 1:1,000), ERβ (Abcam; ab3576; 1:1,000), UBE3A (Proteintech; 10344-1-Ap; 1:1,000), α-Tubulin (Boster; BM1452; 1:1,000), LaminA/C (Proteintech; 10298-1-AP; 1:1,000), MYC-tag (Proteintech; 60003-2-1; 1:1,000), Flag tag (Proteintech; 66008-2-1g; 1:1,000), V5-ub (Proteintech; 14440-1-AP; 1:1,000) and β-actin (CST; 4970; 1:1,000) at 4 °C overnight. And the membranes were incubated with the secondary antibodies. The proteins were detected with ECL chemiluminescent regents and visualized using Tanon 5500 (Tanon Science & Technology; Shanghai, China).
MTT assay
HCT116 and SW620 cells were separately seeded into 96-well plates with five replicates. After the cells had been incubated for the time indicated, 20 mL MTT solution (5 mg/mL) was added to each well. After 4 hours, the solution was removed with a pipette, and 150 mL of dimethyl sulfoxide (DMSO) was added to each well. The plates were shaken gently for 10 minutes. Finally, the OD (optical density) values were detected at 570 nm using a Sunrise microplate reader.
Real-time polymerase chain reaction
Total RNA was extracted from cell lines using TRIzol. Then, reverse transcription of total RNA into cDNA was performed using PrimeScript RT Master Mix (TaKaRa, Tokyo, Japan). To detect mRNA, quantitative real-time polymerase chain reaction (RT-PCR) was conducted using SYBR-green PCR MasterMix. Related primers for qRT-PCR in this research were shown in the Table 1.

Full table
Protein degradation assay
NDRG2-overexpressing (NDRG2+) HCT116 and NDRG2+ SW620 cells, and cells with normal NDRG2 expression (controls) were incubated with CHX (20 µg/mL, Sigma) for the different time (2, 4, or 6 hours) or with MG132 (50 µmol/mL) for 6 hours. Afterward, the cells were harvested, and western blotting (WB) was performed as described above.
Scratch migration assay
Three straight lines were drawn on the back of a six-well plate with a marker pen. Approximately 5×105 cells were seeded into each well. Every well was scratched with medium pipettes perpendicular to the three straight lines. When the cells reached 80–90% confluence, the wells were washed 3 times with phosphate buffer saline (PBS) to remove the loose cells. Free-serum medium was then added to each well. Images were obtained 0–96 hours (0, 24, 48, 72 and 96 hours) later, and the distance from the lines was measured using Photoshop CC2019.
Immunoprecipitation assay
The cells were then harvested with lysis buffer (Beyotime, P0013), and the proteins were quantified using the Pierce BCA Protein Assay Kit (Thermo Scientific, 23225) and adjusted to equal concentrations. Total protein (1 mg) were then immune-precipitated with 1 µL primary ERβ antibody overnight at 4 °C, which had been prebound to magnetic beads. Then, bead-antibody complexes were washed thoroughly with NETN lysis buffer [100 mM NaCl + 20 mM Tris-Cl (pH 8.0) + 0.5 mM EDTA] and resuspended in 1× loading buffer. The samples pulled down with 5% input were resolved on SDS-PAGE gels and transferred to NC membranes. Then, the membranes were detected with appropriate antibodies and visualized using a Tanon 6200 Luminescent Imaging Workstation.
Nuclear and cytoplasmic protein extraction
According to the protocol of a nuclear and cytoplasmic protein extraction kit (Beyotime), cells were lysed with plasma protein extraction reagent A with 1 nmol/L Phenylmethylsulfonyl fluoride (PMSF). After vortexing for 5 seconds, plasma protein extraction reagent B was added to the liquid complex. Then, after high-speed centrifugation, the supernatant was extracted. Nuclear protein extraction reagent was mixed with the sediment, and the mixture was subjected to high-speed vortexing for 30 minutes. Finally, high-speed centrifugation was performed to extract the nuclear protein.
Animals
NDRG2 conditional knockout (NDRG2-/-) mice were supplied by Shanghai Biomodel Organism Science & Technology Development Co., Ltd. All mice in this study were raised under specific-pathogen-free conditions and were aged 8 weeks at the time of the experiments. Wild-type (WT) and NDRG2-/- mice were age-matched and then humanely euthanized. The colon tissues of the mice were harvested, and proteins were extracted from the collected tissue for WB analysis.
Experiments were performed under a project license (No. 81672751) granted by committee ethics board of Xijing Hospital, in compliance with institutional guidelines for the care and use of animals.
Statistical analysis
All data were expressed as mean ± SD. Statistical analyses were conducted using the R software (version 3.63). Differences between two groups were analyzed with Student’s t-test and more than two groups were tested by one-way ANOVA analysis. Differential gene expression in normal and cancerous tissues was analyzed using the Wilcoxon rank-sum test, and the log-rank test was employed to compare survival differences. Statistical significance was considered to exist when P<0.05.
Results
NDRG2 and ERβ were both down-regulated in cancerous tissues of CRC patients
Clinicopathological parameters of 34 patients with CRC are presented in Table 2. IHC (×50, ×200) of cancer tissue samples revealed that all patients with a high expression of NDRG2 also had an increased expression of ERβ (Figure 1A). In addition, three paired WT and NDRG2(-/-) mice colon tissues were selected, and IHC (×200) showed that ERβ expression in the NDRG2(-/-) mice was decreased compared to that in the WT mice (Figure 1B). In addition, there was a positive correlation between the H-scores of NDRG2 and ERβ between the 34 patients (Figure 1C). Analysis of data obtained from the public clinical database TCGA revealed that both NDRG2 and ERβ expression was lower in cancer tissues than in normal tissues (Figure 1D).
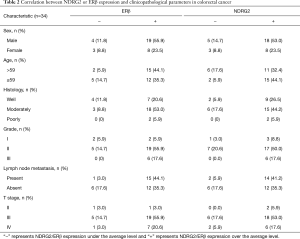
Full table
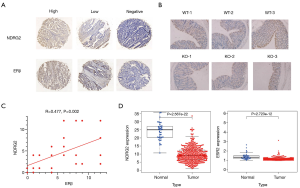
Function analysis and clinical relevance of NDRG2 and ESR2
To validate the biological function of the two genes (NDRG2 and ESR2), GSEA was conducted. The results are shown in Figure 2A,B. The KEGG enrichment analysis showed that NDRG2 was mainly enriched in butanoate metabolism, fatty acid metabolism, oxidative phosphorylation, the pentose phosphate pathway, peroxisome, propanoate metabolism, and pyruvate metabolism. Likewise, ESR2 was mainly enriched in toll-like receptor signaling pathways, apoptosis, the B-cell receptor signaling pathway, the JAK-STAT signaling pathway, inositol phosphate, primary immunodeficiency, and the T-cell receptor signal pathway.
To determine the clinical relevance of NDRG2 and ESR2, we analyzed the association between the differential expression of these two genes and clinical stage. The results indicated that ESR2 expression was negatively associated with stage (P=0.012), whereas NDRG2 expression did not significantly decline as the CRC stage advanced (Figure 2C). To investigate the prognostic value of NDRG2 and ERβ expression, survival analysis of 379 CRC patients was also carried out. Results showed that patients with high NDRG2 and ESR2 (encoding ERβ) expression exhibited a better prognosis than those with low expression (Figure 2D).
NDRG2 improved the protein stability of ERβ at the posttranslational level
To further investigate the relationship between NDRG2 and ERβ, we established NDRG2+ cell lines (HCT116 and SW620). The results of WB and RT-PCR analysis showed that the NDRG2+ group had higher ERβ protein expression than the control group while ERβ RNA expression had no significant alternation in two groups (Figure 3A,B). Compared with those of the WT mice, the colon tissues of the NDRG2(-/-) mice showed a lower level of ERβ (Figure 3A). This phenomenon could be explained by the fact that NDRG2 upregulates ERβ through post-transcriptional activation rather than through transcriptional activation.
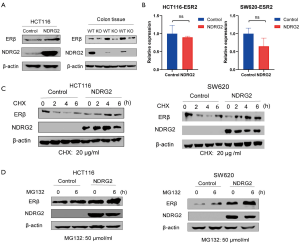
To verify the post-transcription effect of NDRG2 on ERβ, CHX (20 µg/mL) was added to HCT116 and SW620 cells. The results showed that the newborn protein of ERβ significantly decreased in a time-dependent manner after 0, 2, 4, and 6 hours (Figure 3C). ERβ expression in the NDRG2+ group was not obviously reduced compared to that in the control group. Interestingly, after 6 hours, the protein levels of ERβ in SW620 cells showed a slight upward trend. Meanwhile, after the addition of MG132 (50 µmol/mL), the protein expression of ERβ increased gradually, while that of the NDRG2+ group became up-regulated (Figure 3D). Together, these data indicated that NDRG2 might regulate ERβ at the posttranslational level and depends on ubiquitination to inhibit the rate of ERβ protein degradation.
NDRG2 increased the inhibitory effect of DPN on proliferation and migration of CRC cells in vitro
To determine the co-effect of ERβ and NDRG2 on CRC cell proliferation and invasiveness, we performed MTT and scratch migration assays. The results showed a 10–20% decrease in the proliferation of the NDRG2+ group after the addition of various doses of DPN (0, 100, 500, and 1,000 nmol/L) (Figure 4A). However, the NDRG2+ and control groups showed no statistical difference in proliferation in the absence of DPN. Further, the results of the scratch assay indicated a shorter migration distance in the NDRG2+ group than in the control group after exposure to DPN for 0–96 hours (Figure 4B,C). Collectively, these results demonstrated that the combination of NDRG2 and DPN enhances the suppressive effect of ERβ on the proliferation and migration of CRC cells.
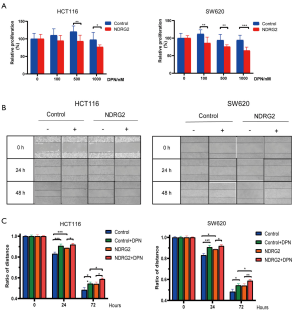
NDRG2 promotes nuclear translocation of ERβ and activates its target genes
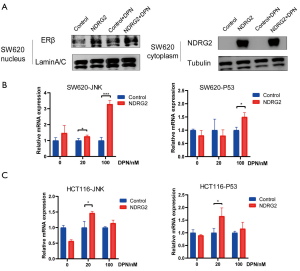
ERs are recognized as nuclear receptors that can dimerize and translocate to the nucleus after binding with ligands and further activate their target genes (17-19). To validate whether NDRG2 promotes the nuclear translocation of ERβ, nuclear pulp separation and RT-PCR analysis were performed. LaminA/C and tubulin are respectively regarded as nuclear and cytoplasmic internal references. The results suggested that NDRG2 could increase the nuclear translocation of ERβ and this effect was enhanced by the addition of DPN (Figure 5A). To explore the potential role of ERβ in nuclear translocation, target genes of ERβ were selected, among which Mitogen-activated protein kinase 8 (MAPK8, also called JNK) and Tumor protein p53 (TP53) were identified as the two most likely target genes. The RT-PCR results showed both JNK and TP53 mRNA expression (Figure 5B,C) to be up-regulated in the NDRG2+ group. Taken together, these results indicated that NDRG2 could facilitate nuclear translocation of ERβ and activate JNK and TP53.
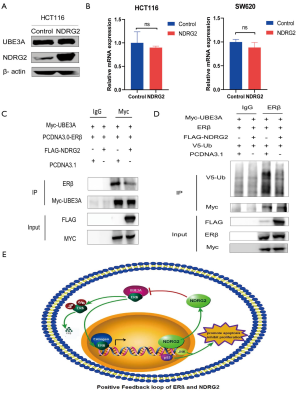
NDRG2 inhibited ubiquitination of ERβ by suppressing ERβ to combine with UBE3A
The above results suggested that NDRG2 might reduce ERβ ubiquitin-associated degradation. To investigate potential mechanism of the effect of NDRG2 on ERβ, we focused on UBE3A in ubiquitin-mediated proteasome degradation. The results showed that NDRG2 had no significant influence on the mRNA or protein expression of UBE3A (Figure 6A,B). Given that NDRG2 can co-locate with ERβ (15) and UBE3A can promote the degradation of ERβ, an immunoprecipitation assay was performed to analyze the underlying role of NDRG2 in UBE3A-ERβ binding. The results demonstrated that NDRG2 could affect the binding of UBE3A to ERβ in 293 T cells (Figure 6C). Moreover, the level of ERβ protein expression retained by UBE3A was significantly lower in the NDRG2+ group. The immunoprecipitation assay was also used to explore whether NDRG2 down-regulates ERβ ubiquitination. Intriguingly, 293T cells transfected with NDRG2 showed a significant reduction in V5-Ub (Figure 6D). Collectively, these results indicated that NDRG2 could suppress the ubiquitination of ERβ by inhibiting the binding of ERβ to UBE3A. In addition, HT29 cells were cultivated and expression of NDRG2 was measured after the addition of 20 µmol/mL DPN. The results showed that ERβ activation could increase the expression levels of NDRG2 in CRC cells (Figure S1).
Discussion
ERβ has been recognized as a tumor suppressor protein (2,4,20-23), and as a positive prognostic indicator for patients with CRC. However, the molecular regulatory network and underlying signaling pathway of ERβ remain largely unknown. In this report, we first demonstrated that NDRG2 reduces ubiquitination-associated degradation of ERβ. Up-regulated ERβ expression was further observed to impose an inhibitory effect on CRC cells. We also saw that UBE3A participates in the ubiquitination of ERβ and NDRG2 blocks UBE3A-ERβ binding to improve ERβ protein stability mechanistically. In the presence of DPN, NDRG2 was also found to promote nuclear translocation of ERβ. In turn, ERβ activates its downstream tumor suppressor genes such as TP53 and JNK (Figure 3B,C). Besides, H-scores of IHC indicated a statistically linear relationship between NDRG2 and ERβ. NDRG2high/ESR2high is a gene signature that might be used as a marker for the prognosis of CRC patients.
NDRG2 and ERβ have been established as pivotal players in the maintenance of colorectal epithelium cell morphology and differentiation (9,21,22). Ma et al. (15) reported that ERβ co-occurs with NDRG2 in astrocytes and DPN could activate ERβ to elevate NDRG2 mRNA expression. Similarly, Li et al. (24) revealed that estrogen interacts with ERβ rather than ERα to up-regulate NDRG2 expression at the transcriptional level. In our study, we demonstrated that ERβ enhances the expression of both NDRG2 mRNA and protein in CRC cells and that NDRG2+ cells showed a higher level of ERβ protein expression. To investigate whether the stabilizing effect of NDRG2 on ERβ is dependent on the regulation of Ub, cells were treated with different doses of CHX and MG132, and the results showed a higher level of ERβ in the NDRG2+ group than the control group. ERs are nuclear receptors that can be transferred into the nucleus and regulate target genes by recognizing estrogen response elements (17-19). We extracted nuclear protein and found that ERβ expression was higher in the NDRG2+ group than in the control group, suggesting that NDRG2 could promote ERβ to translocate into the nucleus and to bind to ERE. It has been reported that via the activation functional domains AF-1 (25) and AF-2 (26), ERs are able to recruit different cofactors to the promoter region of target genes. A growing bank of research has revealed that through various mechanisms (27), ERβ can induce apoptosis and reduce proliferation. These mechanisms include DNA fragmentation (27), TP53 signaling enhancement (28), JNK pathway amplification (29), and cellular differentiation (20,30). Also, Kao et al. have shown that ERβ-UBE3A interaction promotes liver cell differentiation while ERα facilitates proliferation (31). However, a previous study showed that without estrogen, MG132 failed to increase basal ERβ-mediated transcription in 293T cells (32). The reasons for the variation in results might include amplification of the effect of NDRG2 on ERβ by DPN or estrogen, and the use of different cell lines in experiments.
UBE3A, which is classified as a member of the functionally related E3-ubiquitin-protein ligase family, is a 100-kDa cellular protein that regulates substrate specificity of the ubiquitin-proteasome degradation system (33). Growing evidence has shown that the degradation of ERs depends on the 26S proteasome pathway (34,35) and is closely related to the transcriptional process (36). A recent study reported that UBE3A could decrease ERβ at the cellular level (36). Likewise, our experiments demonstrated that UBE3A could degrade ERβ in a Ub-dependent manner and that NDRG2 could inhibit the binding of UBE3A to ERβ, thus reducing ERβ ubiquitination.
Limitations of the present study include the relatively small sample size and inadequate elucidation of the regulatory network between NDRG2 and ERβ. Further studies to address these issues are needed.
In the current study, we demonstrated for the first time that NDRG2 could reduce ERβ degradation and enhance its tumor-suppressive effect by inhibiting UBE3A-ERβ binding. Given that ERβ could transactivate NDRG2 and apoptosis-related genes, we discovered that there is a positive feedback loop between NDRG2 and ERβ (Figure 6E), which can promote the differentiation and inhibit the proliferation of CRC cells. Furthermore, the NDRG2high/ESR2high signature is associated with a good clinical outcome in CRC patients and could be used as a prognostic marker of CRC.
Acknowledgments
Funding: This work was supported in part by grants from the National Natural Science Foundation of China (81672751) and the Key Research and Development Program of Shaanxi (2019SF-010).
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/jgo-20-557
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jgo-20-557
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jgo-20-557). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Xijing Hospital Ethics Committee (No. KY20203269-1) and informed consent was taken from all the patients. Animal experiments were performed under a project license (No. 81672751) granted by committee ethics board of Xijing Hospital, in compliance with institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Dawood S. The evolving role of immune oncology in colorectal cancer. Chin Clin Oncol 2018;7:17. [Crossref] [PubMed]
- McCashland T. M, Brand R, Lyden E, et al. Gender differences in colorectal polyps and tumors. Am J Gastroenterol 2001;96:882-6. [Crossref] [PubMed]
- Cotrim CZ, Fabris V, Doria ML, et al. Estrogen receptor beta growth-inhibitory effects are repressed through activation of MAPK and PI3K signalling in mammary epithelial and breast cancer cells. Oncogene 2013;32:2390-402. [Crossref] [PubMed]
- Sun ZH, Hu Y, Wang M, et al. Estrogen receptor β and estrogen receptor α36 predict differential outcome of patients with breast cancer. Transl Cancer Res 2018;7:363-70. [Crossref]
- Omoto Y, Iwase H. Clinical significance of estrogen receptor β in breast and prostate cancer from biological aspects. Cancer Sci 2015;106:337-43. [Crossref] [PubMed]
- Konstantinopoulos PA, Kominea A, Vandoros G, et al. Oestrogen receptor beta (ERbeta) is abundantly expressed in normal colonic mucosa, but declines in colon adenocarcinoma paralleling the tumour's dedifferentiation. Eur J Cancer 2003;39:1251-8. [Crossref] [PubMed]
- Jassam N, Bell SM, Speirs V, et al. Loss of expression of oestrogen receptor beta in colon cancer and its association with Dukes' staging. Oncol Rep 2005;14:17-21. [PubMed]
- Kim YJ, Yoon SY, Kim JT, et al. NDRG2 expression decreases with tumor stages and regulates TCF/beta-catenin signaling in human colon carcinoma. Carcinogenesis 2009;30:598-605. [Crossref] [PubMed]
- Assämäki R, Sarlomo-Rikala M, Lopez-Guerrero JA, et al. Array comparative genomic hybridization analysis of chromosomal imbalances and their target genes in gastrointestinal stromal tumors. Genes Chromosomes Cancer 2007;46:564-76. [Crossref] [PubMed]
- Liu N, Wang L, Liu X, et al. Promoter methylation, mutation, and genomic deletion are involved in the decreased NDRG2 expression levels in several cancer cell lines. Biochem Biophys Res Commun 2007;358:164-9. [Crossref] [PubMed]
- Hu XL, Liu XP, Lin SX, et al. NDRG2 expression and mutation in human liver and pancreatic cancers. World J Gastroenterol 2004;10:3518-21. [Crossref] [PubMed]
- Chu D, Zhang Z, Li Y, et al. Prediction of colorectal cancer relapse and prognosis by tissue mRNA levels of NDRG2. Mol Cancer Ther 2011;10:47-56. [Crossref] [PubMed]
- Chen W, Peng J, Ou Q, et al. Expression of NDRG2 in Human Colorectal Cancer and its Association with Prognosis. J Cancer 2019;10:3373-80. [Crossref] [PubMed]
- Ma YL, Qin P, Feng DY, et al. Estrogen regulates the expression of Ndrg2 in astrocytes. Brain Res 2014;1569:1-8. [Crossref] [PubMed]
- Shen L, Qu X, Li H, et al. NDRG2 facilitates colorectal cancer differentiation through the regulation of Skp2-p21/p27 axis. Oncogene 2018;37:1759-74. [Crossref] [PubMed]
- Jonsson P, Katchy A, Williams C. Support of a bi-faceted role of estrogen receptor β (ERβ) in ERα-positive breast cancer cells. Endocr Relat Cancer 2014;21:143-60. [Crossref] [PubMed]
- Ascenzi P, Bocedi A, Marino M. Structure-function relationship of estrogen receptor alpha and beta: impact on human health. Mol Aspects Med 2006;27:299-402. [Crossref] [PubMed]
- Chang EC, Frasor J, Komm B, et al. Impact of estrogen receptor beta on gene networks regulated by estrogen receptor alpha in breast cancer cells. Endocrinology 2006;147:4831-42. [Crossref] [PubMed]
- Schleipen B, Hertrampf T, Fritzemeier KH, et al. ERβ-specific agonists and genistein inhibit proliferation and induce apoptosis in the large and small intestine. Carcinogenesis 2011;32:1675-83. [Crossref] [PubMed]
- Wada-Hiraike O, Imamov O, Hiraike H, et al. Role of estrogen receptor beta in colonic epithelium. Proc Natl Acad Sci U S A 2006;103:2959-64. [Crossref] [PubMed]
- Giroux V, Lemay F, Bernatchez G, et al. Estrogen receptor beta deficiency enhances small intestinal tumorigenesis in ApcMin/+ mice. Int J Cancer 2008;123:303-11. [Crossref] [PubMed]
- Hartman J, Edvardsson K, Lindberg K, et al. Tumor repressive functions of estrogen receptor beta in SW480 colon cancer cells. Cancer Res 2009;69:6100-6. [Crossref] [PubMed]
- Li Y, Yang J, Li S, et al. N-myc downstream-regulated gene 2, a novel estrogen-targeted gene, is involved in the regulation of Na+/K+-ATPase. J Biol Chem 2011;286:32289-99. [Crossref] [PubMed]
- Paech K, Webb P, Kuiper GG, et al. Differential ligand activation of estrogen receptors ERalpha and ERbeta at AP1 sites. Science 1997;277:1508-10. [Crossref] [PubMed]
- Benecke A, Chambon P, Gronemeyer H. Synergy between estrogen receptor alpha activation functions AF1 and AF2 mediated by transcription intermediary factor TIF2. EMBO Rep 2000;1:151-7. [Crossref] [PubMed]
- Qiu Y, Waters CE, Lewis AE, et al. Oestrogen-induced apoptosis in colonocytes expressing oestrogen receptor beta. J Endocrinol 2002;174:369-77. [Crossref] [PubMed]
- Hsu HH, Cheng SF, Wu CC, et al. Apoptotic effects of over-expressed estrogen receptor-beta on LoVo colon cancer cell is mediated by p53 signalings in a ligand-dependent manner. Chin J Physiol 2006;49:110-6. [PubMed]
- Razandi M, Pedram A, Greene GL, et al. Cell membrane and nuclear estrogen receptors (ERs) originate from a single transcript: studies of ERalpha and ERbeta expressed in Chinese hamster ovary cells. Mol Endocrinol 1999;13:307-19. [Crossref] [PubMed]
- Wada-Hiraike O, Warner M, Gustafsson JA. New developments in oestrogen signalling in colonic epithelium. Biochem Soc Trans 2006;34:1114-6. [Crossref] [PubMed]
- Kao TL, Kuan YP, Cheng WC, et al. Estrogen receptors orchestrate cell growth and differentiation to facilitate liver regeneration. Theranostics 2018;8:2672-82. [Crossref] [PubMed]
- Pal P, Lochab S, Kanaujiya JK, et al. E6AP, an E3 ubiquitin ligase negatively regulates granulopoiesis by targeting transcription factor C/EBPα for ubiquitin-mediated proteasome degradation. Cell Death Dis 2013;4:e590. [Crossref] [PubMed]
- Huibregtse JM, Scheffner M, Beaudenon S, et al. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci U S A 1995;92:5249. [Crossref] [PubMed]
- Nawaz Z, Lonard DM, Dennis AP, et al. Proteasome-dependent degradation of the human estrogen receptor. Proc Natl Acad Sci U S A 1999;96:1858-62. [Crossref] [PubMed]
- Marsaud V, Gougelet A, Maillard S, et al. Various phosphorylation pathways, depending on agonist and antagonist binding to endogenous estrogen receptor alpha (ERalpha), differentially affect ERalpha extractability, proteasome-mediated stability, and transcriptional activity in human breast cancer cells. Mol Endocrinol 2003;17:2013-27. [Crossref] [PubMed]
- Picard N, Charbonneau C, Sanchez M, et al. Phosphorylation of activation function-1 regulates proteasome-dependent nuclear mobility and E6-associated protein ubiquitin ligase recruitment to the estrogen receptor beta. Mol Endocrinol 2008;22:317-30. [Crossref] [PubMed]
(English Language Editor: J. Reynolds)


