
Increased expression of MCM4 is associated with poor prognosis in patients with hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is one of the most common causes of cancer-related deaths. A million liver cancer incidences and 829,000 associated deaths occurred in 2016, worldwide (1). Advances in the early detection and treatment of liver cancer have improved the overall survival (OS) rate of patients; however, it continues to be responsible for many cancer-related mortalities. Hence, to improve the OS of liver cancer patients, there is a pressing need to find novel therapeutic targets. The replication of DNA is of specific interest during the study of tumor formation and tumorigenesis. Complete and accurate DNA replication is the foundation for cell proliferation and genomic stability (2). Genes that drive cell proliferation induce DNA replication stress, leading to genomic instability and selection to avoid apoptosis (3). The minichromosome maintenance complex components (MCMs) family, a replicative DNA helicase that forms a hexameric ring-shaped complex around the DNA, plays a central role in the DNA replication process. The MCM2–7 complex possesses helicase activity and is involved in the formation of the pre-replication complex, which helps in the unwinding of DNA, leading to the recruitment of DNA polymerase, and initiation of DNA replication followed by extension (4,5). Accumulating evidences have shown that MCMs can predict the progression of tumors and are correlated with the prognosis of cancer patients. It has been reported that MCM2 is a useful screening marker in cervical, esophageal, and colorectal cancers (6-8). It has been reported that MCM3 is a new proliferation marker in oral squamous cell carcinoma (9). The G364R mutation of minichromosome maintenance complex component 4 (MCM4), detected in human skin cancer cells, has been known to affect the DNA helicase activity of the MCM4/6/7 complex (10). It has been reported that MCM5 is a prognostic biomarker in breast cancer (11). Overexpression of MCM6 indicates a low survival rate in glioma patients (12). Additionally, MCM7 was identified as a predictor of pancreatic neuroendocrine tumor progression and a biomarker for the early screening of gastric cancer (13,14).
Furthermore, there are reports on the regulatory expression of individual MCMs in HCC. As a novel marker of cell dynamic index, MCM2 can be used to evaluate the progression from cirrhosis to HCC (15,16). As a possible novel target for the treatment of liver cancer, MCM3 has been reported to be associated with the resistance of liver cancers (17). The MCM5 is one of the seven senescence-associated gene (SAG) signatures, which can predict the OS of Asian patients with liver cancer (18). The MCM6 has been known to promote metastasis of HCC through the mitogen-activated protein kinase kinase/extracellular-signal-regulated kinase (MEK/ERK) pathway and can be used as a novel serum marker for the diagnosis of early recurrence of HCC (19). The MCM7 promotes tumor progression through cyclin D1-dependent signals and serves as a prognostic marker for HCC patients (20). Furthermore, the expression of MCM7 can predict the prognosis of HCC, post-surgery (21). However, there was few studies about the effects of MCM4 on HCC. The prognostic importance of MCM4 in HCC remains to be explored.
Given the high expression of MCM4 in multiple HCC-related expression datasets, in the current study, we used a large number of public expression and survival datasets collected from different online platforms to comprehensively analyze the expression pattern and clinical significance of MCM4 in HCC patients. Furthermore, we analyzed the genes that were co-expressed with MCM4 in HCC. These genes may be associated with the underlying mechanisms of MCM4-related progression and prognosis of HCC. Our results may provide evidence for using MCM4 as a potential prognostic biomarker for liver cancer treatment.
We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/jgo-20-574).
Methods
Expression of the MCM4 gene in different cancers
The expression level of MCM4 mRNA in different tumor and normal tissues was analyzed using the following databases: Oncomine (https://www.oncomine.org/resource/login.html) (22,23), Gene Expression Profiling Interactive Analysis 2 (GEPIA2; http://gepia2.cancer-pku.cn) (24), and the Gene Expression database of Normal and Tumor tissues (GENT; http://gent2.appex.kr/gent2) (25). In GEPIA2, the MCM4 expression in tumor samples from The Cancer Genome Atlas (TCGA) was compared with that in the normal adjacent tissues from TCGA as well as normal healthy liver tissues from the Genotype-Tissue Expression (GTEx) database (26). We queried MCM4 in the databases to obtain the respective expression patterns.
Expression of MCM4 gene and protein in liver cancer
The MCM4 expression in HCC and normal tissues was obtained from the Oncomine, HCCDB (https://bigd.big.ac.cn/databasecommons/database/id/6024) (27), and UALCAN (http://ualcan.path.uab.edu/index.html) (28) databases. In the Oncomine, by using the following parameter thresholds (I) fold-change (FC) =2, (II) gene rank 10%, and (III) P value =1E−4, we obtained the FCs of MCM4 mRNA in liver cancer compared to those in the normal liver tissues. Next, to evaluate the relative expression of MCM4 in liver cancer and normal liver tissues, the standardized original transcriptome data were analyzed. The MCM4 protein expression in liver cancer and normal liver tissues was obtained based on the immunohistochemistry (IHC) results retrieved from the Human Protein Atlas (HPA; https://www.proteinatlas.org) database (29).
Expression of MCM4 gene at mRNA and protein levels in liver cancer
HCC tissues (n=30) and adjacent normal tissues (n=30) were collected from patients who diagnosed with HCC in Affiliated Dongguan People’s Hospital, Southern Medical University. All tissues obtained from surgery process were placed in liquid nitrogen and stored at -80°C. Participators who received any chemotherapy and radiation treatment prior to study were except in our study. Their TNM stages included I and IV. Written informed consent was obtained prior to study. This study was approved by the Ethic Committee of Affiliated Dongguan People’s Hospital, Southern Medical University. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Expression of MCM4 was assessed by RT-qPCR
Total RNA was extracted from HCC tissues using RNAsimple Total RNA Kit (Tiangen, Beijing, China). Thereafter, RT and qPCR were conducted using FastKing Real Time One Step RT-qPCR (SYBR Green) kit (Tiangen). The reaction of RT was 50 °C for 30 min, and qPCR reactions were 95 °C for 3 min (pre-degeneration), 40 cycles of 95 °C for 15 sec (degeneration) and 60 °C for 30 sec (annealing and extending) on CXF96 real time PCR system (BioRad). The relative expression was computed by 2−ΔΔCt method.
Expression of MCM4 was examined by western blot
The HCC and adjacent normal tissues were lysed using RIPA lysis buffer (MCE, Shanghai, China) to extract total protein. After measuring the protein concentration using BCA Protein Assay Kit (Sangon, Shanghai, China), 50 µg protein were separated by 12% SDS-PAGE. The separated proteins were transferred onto PVDF membranes and blocking using 5% skim milk. The membranes were incubated with anti-MCM4 (#3228, 1:1,000; Cell Signaling Technology) and anti-GAPDH (#2118, 1:1,000; Cell Signaling Technology) overnight at 4 °C and incubated with HRP-conjugated Goat Anti-Rabbit IgG (D110058, 1:10,000) (Sangon). The protein signals were visualized using ECL luminescence reagent (Sangon).
Expression of MCM4 was evaluated by IHC staining
Tissue sections were dewaxed with xylene and rehydrated with gradient alcohol. After incubation in Hydrogen Peroxide Block for 15 min, the sections were blocking with goat serum. Primary anti-MCM4 (#12973, 1:200; Cell Signaling Technology) was added to incubate with the sections at 4 °C overnight and HRP-conjugated Goat Anti-Rabbit IgG (D110058, 1:10,000) (Sangon) was added for incubation for 30 min at room temperature. After DAB staining and hematoxylin counterstaining, the sections were visualized by a digital microscope.
Relationship between MCM4 expression and clinical characteristics of liver cancer patients
The UALCAN web portal, which uses TCGA data, was used with default settings to verify the mRNA expression and promoter methylation of MCM4 associated with various characteristics of HCC patients (28). The MCM4 expression in tumor tissues was compared with that in normal liver tissues based on the following patient characteristics: sample type, stage, age, race, gender, and grade. Further, we analyzed the pattern of MCM4 expression in various clinicopathological characteristics of HCC patients using TCGA data from the University of California, Santa Cruz (UCHCC) Xena platform (http://xena.ucHCC.edu) (30,31). The promoter methylation was analyzed in light of the stage, age, race, gender, and grade of HCC patients. The data for promoter methylation corresponded to the following probes on the 450K human methylation chip of Infinium in the UALCAN web: cg11882144, cg24792272, cg24563117, cg17660110, cg00318618, cg09303136, cg07417733, and cg22691796.
MCM4 gene mutations and copy number alterations (CNAs) in HCC
We used the cBioPortal (http://www.cbioportal.org) (32,33) to analyze the mutations and CNAs of the MCM4 gene, in HCC. The location and frequency of mutations were identified in five hepatoma studies from the cBioPortal. The CNAs were obtained from RNA-sequencing (RNA-seq) data through the genomic identification of significant targets in cancer (GISTIC) algorithm (default settings), and mRNA expression data was drawn from the cBioPortal network. The software GraphPad Prism 7.0 (GraphPad software, San Diego, CA, USA) was used for statistical analysis.
Correlation between MCM4 expression and survival of HCC patients
Using the Kaplan-Meier plotter (http://kmplot.com/analysis) (34), the clinical data from TCGA, Gene Expression Omnibus (GEO), and t European Genome-phenome Archive were correlated with the mRNA expression level of MCM4 to study the clinical significance between its expression and liver cancer. The platform was used to determine the prognosis of the disease, including OS, relapse-free survival (RFS), progression-free survival (PFS), and disease-specific survival (DSS). The expression of MCM4 in each group was drawn into a survival curve, and the prognosis exponent of each sample was assessed by Cox survival analysis. We used the data from all participants or clinicopathological subgroups to automatically select the best cut-off mode, which was used to divide the queue to generate the survival curve. All the other default settings were preserved. Further, univariate and multivariate survival analyses were performed using the Kaplan-Meier plotter. Statistical significance was estimated by the Cox regression tests, the Kruskal-Wallis test, or the Mantel-Cox log-rank test. The relevance of MCM4 expression in the survival of patients was also investigated in HCC and adjacent normal tissues using the HCCDB (https://bigd.big.ac.cn/databasecommons/database/id/6024) (27).
Analysis of MCM4-co-expressed genes in HCC
The co-expression genes associated with MCM4 in HCC were derived from the Oncomine database. Further, we used the UCHCC Xena platform (30,31) to create a heat map of the co-expressed genes using the HCC data from TCGA, and confirmed the correlation between the MCM4 and the co-expressed genes.
Gene Ontology and signaling pathway analyses of MCM4-co-expressed genes
The pathway [the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Reactome] and Gene Ontology (GO) analyses of MCM4-co-expressed genes (obtained from the Oncomine database) were performed using the NetworkAnalyst 3.0 tool (https://www.networkanalyst.ca) (35). The enriched GO annotations and pathways were visualized as network diagrams.
Statistical analysis
The data were analyzed by GraphPad 6.0 and presented as mean ± SD. The paired Student’s t-test was used to analyze the differences between paired HCC tissues and adjacent normal tissues. Additionally, the Student’s t-test was used to analyze the differences between other two variables except tissues, whereas one-way ANOVA was used to analyze differences across > two variables. P value was shown in the bar chat. P<0.05 was considered to have significant difference.
Results
MCM4 gene expression in different cancers
To explore MCM4 expression patterns in different cancer types versus normal tissues, we used three bioinformatics databases. In the Oncomine, a great many distinct analyses showed differences in the MCM4 expression between the cancer and normal tissues. Further, our results showed that the expression of MCM4 was found to be upregulated in the following cancers compared to their corresponding normal tissues: bladder, cervical, colorectal, head and neck, lung, ovarian, pancreatic, melanoma, stomach, prostate, sarcoma, lymphoma, and esophageal carcinoma (Figure 1A). The expression of MCM4 was also found to be higher in HCC compared to the normal liver tissue. Furthermore, only the kidney cancer showed a downregulation of MCM4 (Figure 1A). Breast cancer, brain, and central nervous system (CNS)-related cancers, and leukemia showed both upregulation and downregulation of MCM4 (Figure 1A). The GEPIA2 database, which integrates expression data from the TCGA and GTEx databases, was used to further analyze the expression of MCM4 in 33 human tumors and their corresponding normal tissues (Figure 1B). Among the 33 cancer types analyzed, all showed higher expressions of MCM4 than the corresponding normal tissues; meanwhile, the MCM4 expression was significantly higher in 20 of them. In the GENT, MCM4 was found to be upregulated in the following cancer types compared to the corresponding normal tissues, based on the expression profile from the U133Plus2.0 microarray platform (GPL570) (Thermo Fisher, Waltham, MA, USA): adrenal gland, bladder, blood, bone, brain, breast, colon, cervix, endometrium, lung, head and neck, kidney, liver, pancreatic, stomach, ovarian, prostate, skin, thyroid, tongue, oral, and uterine cancers (Figure 1C). The expression level of MCM4 was found to be higher in HCC than that in normal tissues (Figure 1C).
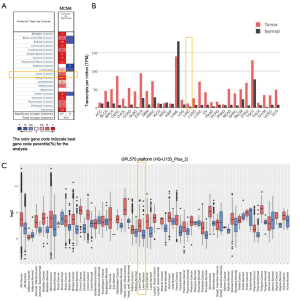
Expression of the MCM4 gene and protein in liver cancer
The expression of the MCM4 gene in HCC was evaluated in several published HCC-related studies using the HCCDB. Analysis of 11 HCC cohorts in the HCCDB showed that the MCM4 mRNA expression of MCM4 was significantly higher in HCC tissues than in adjacent normal tissues (Figure 2A,B). Similarly, results from the Oncomine database indicated a higher expression of MCM4 in HCC tissues than in adjacent normal tissues in the following datasets: ‘Roessler Liver’, ‘Roessler Liver 2’, and ‘Wurmbach Liver’ (Figure 2C). The upregulation of MCM4 in HCC compared to the normal liver tissues was further confirmed by the UALCAN database, which is based on TCGA data (Figure 2D). Additionally, the expression of MCM4 was verified at the protein level in normal hepatocytes and HCC tissues using IHC data obtained from the HPA project. The results showed that two of the 11 liver cancer patients had weak to moderate staining, whereas normal healthy liver cells had no detectable staining, indicating no MCM4 expression in healthy liver tissues (Figure 2E). Overall, multiple databases showed that MCM4 is upregulated in HCC compared to the normal liver tissues, both at the mRNA and protein level.
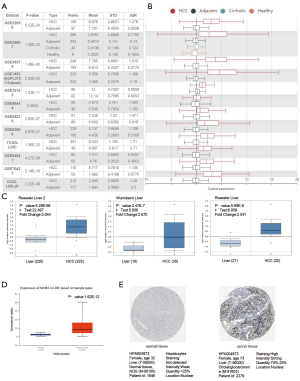
Expression of MCM4 gene at mRNA and protein levels in liver cancer
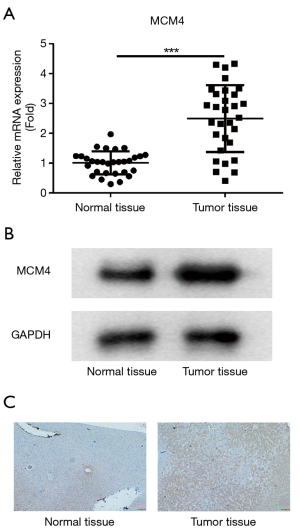
To confirm the results showed above, mRNA expression of MCM4 was measured by RT-qPCR. The results demonstrated that MCM4 expression was upregulated in tumor tissues, compared with that in normal tissues (Figure 3A). Furthermore, expression of MCM4 at protein level was tested by western blot, and the data showed that MCM4 in tumor tissues was higher than normal tissues (Figure 3B). On the other hand, the results indicated that protein MCM4 level was increased in HCC tissues, which was assessed by IHC (Figure 3C).
Relationship between MCM4 expression and clinicopathological characteristics of liver cancer patients
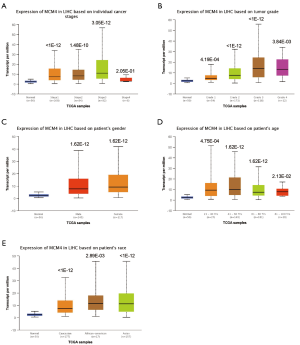
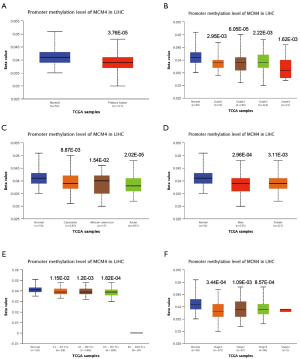
We used the UALCAN and UCHCC Xena platforms to study the relationship between MCM4 mRNA expression and the clinicopathological characteristics of liver cancer patients, based on TCGA data. Compared to the normal tissues, MCM4 expression was significantly upregulated in HCC, irrespective of stage, grade, gender, age, or race (Figure 4A,B,C,D,E, Table 1). Other clinicopathological characteristics, including surgical intervention, drug therapy, radiotherapy, liver transplantation, and ablation, exhibited no significant correlation with MCM4 expression in patients with liver cancer (Table 1). Furthermore, promoter methylation serves as an important epigenetic regulator of gene expression. In the UALCAN analysis of TCGA data, normal methylation of HCC tumors was significantly reduced compared with that of the normal tissues (Figure 5A). Additionally, promoter methylation levels were reduced despite patient characteristics, including stage, grade, gender, age, and race (Figure 5B,C,D,E,F). Hence, MCM4 expression may be inversely correlated with methylation of certain CpG sites. Together, these data indicate an increase in mRNA expression and a decrease in promoter methylation of MCM4 in HCC.
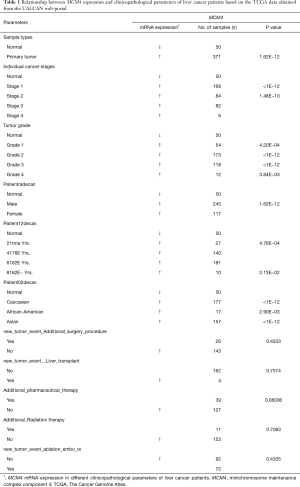
Full table
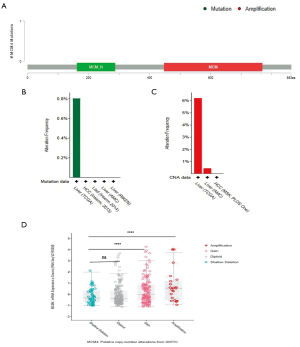
MCM4 gene mutations and CNAs in HCC
We used the cBioPortal web to analyze mutations and CNAs associated with the MCM4 gene in a group of HCC patients. Three mutations were found in the MCM4 protein, most of which were located in the MCM domain (Figure 6A). In addition, the mutation frequency, as shown by the TCGA dataset, was approximately 0.8% (Figure 6B). These results showed that CNAs were more common in MCM4. In the CNAs, amplification was the primary alteration, observed in ~6% of the patients (Figure 6C). Thus, a significant correlation was noted between CNAs in HCC and MCM4 expression, as indicated by the TCGA data (Figure 6D). Most notably, amplification and gain were related to the MCM4 expression. These data suggest that the increase in MCM4 expression in HCC may be partly due to the CNAs.
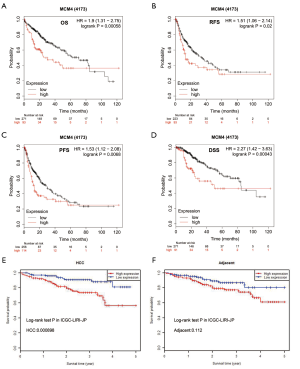
Correlation between MCM4 expression and survival of patients with HCC
Although MCM4 plays an important role in the carcinogenesis process in humans, the relationship between MCM4 expression and clinical prognosis is not clear. In the current study, the Kaplan-Meier analysis showed the relationship between MCM4 expression and the survival of liver cancer patients with different clinicopathological characteristics (Figure 7). Specifically, the OS, RFS, PFS, and DSS were shown to be negatively related with MCM4 expression in HCC patients (Figure 7A,B,C,D). In addition, the Kaplan-Meier plotter was used to perform univariate and multivariate analyses between MCM4 expression and various clinicopathological parameters. The univariate analysis showed that MCM4 mRNA expression was significantly correlated with the OS of HCC patients with different clinicopathological features, including male, Asian, grade 3, stages T2 and T3, stages 2 and 3, no vascular invasion of HCC, no alcohol consumption, and no hepatitis virus infection (Table 2). The multivariate analysis also showed that there was a significant correlation between the expression of MCM4 and the OS rate of patients with liver cancer (Table 2). Log-rank test analysis showed the relationship between MCM4 expression in tumor or adjacent normal tissues and the survival of patients with HCC (Figure 7). The OS of patients was significantly, negatively correlated with the MCM4 expression in HCC tissues, but not in adjacent normal tissues (Figure 7E,F). These analyses confirmed the correlation between MCM4 expression and the prognosis of patients with liver cancer. Data were analyzed using the Kaplan-Meier Plotter network.
Full table
Analysis of MCM4-co-expressed genes in HCC
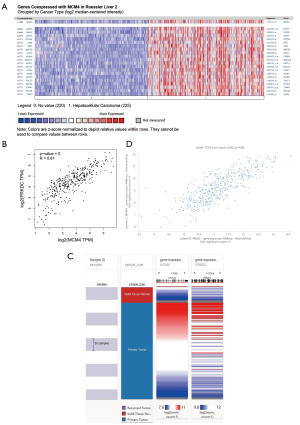
We used Oncomine (Cho HCC dataset) to study the genes co-expressed with MCM4 in HCC. As shown in Figure 8A, we identified a group of MCM4-co-expressed genes in HCC. Among these, PRKDC (protein kinase, DNA-activated, catalytic subunit) was found to be the most highly co-expressed (r=0.846). Further, analysis of TCGA data using GEPIA2 confirmed that MCM4 was indeed positively correlated with the expression of PRKDC (Figure 8B). Ultimately, the Pearson and Spearman rank correlation analyses confirmed that there was a positive correlation between these two genes in HCC patients based on the TCGA data from the UCHCC Xena web platform (Figure 8C,D). Our findings suggest that MCM4 and PRKDC expression may be closely related and may be involved in the signaling pathways related to HCC.
Analysis of MCM4-co-expressed genes revealed liver cancer-related signaling pathways
To identify the signaling pathways associated with MCM4 in HCC, 17 of the MCM4-co-expressed genes (including MCM4) originated from the Oncomine database (Figure 7A) were analyzed. The top ten pathways originated from the Reactome database for the MCM4-correlated genes were ‘mitotic prometaphase’, ‘resolution of sister chromatid cohesion’, ‘cell cycle’, ‘M phase’, ‘mitotic M-M/G1 phases’, ‘separation of sister chromatids’, ‘mitotic anaphase’, ‘mitotic metaphase and anaphase’, ‘cell cycle, mitotic’, and ‘deposition of new CENPA-containing nucleosomes at the centromere’ (Figure 9A). The top four KEGG pathways associated with the MCM4 positively correlated genes were ‘cell cycle’, ‘progesterone-mediated oocyte maturation’, ‘oocyte meiosis’, and ‘non-homologous end-joining’ (Figure 9B). These pathways may be associated with the development and progression of liver cancer.
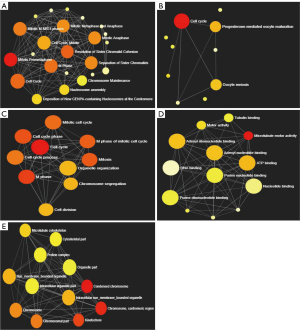
Furthermore, we used NetworkAnalyst 3.0 (http://www.networkanalyst.ca) to analyze the GO annotations, such as biological processes (BP), molecular functions (MF), and cellular components (CC), associated with MCM4 and the co-expressed genes. We found that MCM4 and the positively correlated genes were the most significantly enriched for ‘cell cycle’ under BP (Figure 9C), ‘microtubule motor activity’ under MF (Figure 9D), and ‘condensed chromosome’ under CC (Figure 9E).
Discussion
Liver cancer is one of the major causes of cancer-related deaths worldwide (2). Early stages of liver cancer are often treatable, with surgical interventions having the highest rate of cure (36). However, despite the use of surgery, interventional therapy, chemotherapy, targeted therapy, and immunotherapy, the mortality rate of liver cancer patients in the late stage of the disease continues to be very high. Recent treatment methods, including targeted therapy based on the liver cancer molecular profile, have improved the survival rate of patients with advanced liver cancer (36). In the current study, the increased MCM4 expression was found to be negatively correlated with the survival rate of HCC patients, suggesting that MCM4 may be used as a therapeutic target for HCC.
The increased expression of MCM4, a member of the MCM family, is reportedly associated with the development and pathological stages of esophageal cancer; thus, it may be a valuable molecular marker (37). Further, the expression of MCM4 is known to be associated with metastasis of melanoma, and may be related to the molecular mechanisms underlying poor prognosis in melanoma patients (38). The role of MCM4 has also been investigated in the proliferation of some non-small-cell lung carcinoma (NSCLC) cells. The high expression of MCM4 in NSCLC and its correlation with non-adenocarcinoma histology indicate that MCM4 may be a used as a therapeutic target in certain subtypes of NSCLC (39). Gene knockout experiments with small interfering (si)RNAs showed that the downregulation of MCM4 inhibited the growth, migration, and invasion of lung adenocarcinoma cells (40). However, the role of MCM4 in the progression of liver cancer has not been explored to date.
In the current study, the prognostic value of MCM4 in liver cancer was identified for the first time through systematic bioinformatics analysis using open-source resources/datasets. Analysis of the datasets from the GEO, TCGA, and HCCDB databases showed that MCM4 was significantly upregulated in HCC tissues, and was negatively correlated with the OS, PFS, RFS, and DSS in HCC patients. Our results also confirmed this trend at the protein level in liver cancer patients. Through the experiments of RT-qPCR, western blot and IHC staining, we verified the level of MCM4 was increased in HCC tissues. Furthermore, univariate and multivariate analyses showed that the expression of MCM4 was significantly correlated with the clinicopathological characteristics of HCC patients. The change in the MCM4 gene expression may be attributed to the epigenetic control, mutations, and CNAs control in cancer cells. Further analysis of TCGA datasets showed that the mutation rate of MCM4 in HCC patients was 0.8%, the amplification rate was 6%, and its methylation was significantly reduced. Promoter methylation was analyzed using TCGA datasets through the UALCAN web portal. We observed a significant difference in the promoter methylation (P=3.76E–05) between the normal liver samples (n=50) and liver cancer samples (n=377). Overall, our results suggest that the increased expression of MCM4 in HCC may be partly caused by one or a combination of these factors. However, this needs to be verified in additional data with more normal control samples and through the use of different methods. Further research is needed to explore the molecular mechanisms associated with MCM4 in the prognosis of HCC patients.
Aiming to explore the changes in various pathways and functions related to MCM4 in liver cancer, we analyzed the genes that were identified as co-expressed with MCM4. The expression of PRKDC was found to be highly correlated with that of MCM4. The correlation between PRKDC and the MCM4 expression patterns was further confirmed by an additional analysis using different online platforms. A member of the phosphatidylinositol (PI)3/PI4 kinase family, PRKDC is involved in DNA double-strand break repair and recombination with the Ku70/Ku80 heterodimer proteins (41). The IHC results obtained from the HPA database showed that >90% of the liver cancer cells expressed the PRKDC protein (unpublished data). Multiple studies have evaluated the role of PRKDC in different cancers.
We analyzed the pathways associated with 17 genes co-expressed with MCM4 to determine the functional importance of MCM4 expression. We used the NetworkAnalyst 3.0 tool (http://www.networkanalyst.ca) to investigate the network of pathways associated with the genes co-expressed with MCM4 in HCC. Moreover, GO enrichment analysis was performed to explore the BP, CC, and MF associated with the co-expressed genes. The results indicated that the co-expressed genes were significantly enriched for cell cycle-related pathways, which has also been found to be important in eribulin chemotherapy in ovarian cancer (42). Eribulin can inhibit the development of ovarian cancer by downregulating the establishment of sister chromatid cohesion N-acetyltransferase (ESCO1) and MCM4 genes, which highlights the potential application of eribulin in inducing cell cycle arrest in ovarian cancer cells. Therefore, MCM4 expression may be a potential biomarker of the therapeutic effect of eribulin on ovarian cancer (42). Association of MCM4 and a group of co-expressed genes in cell cycle pathway-related modules suggests the potential role of MCM4 in tumor chemotherapy. The GO analysis indicated that the two most abundant CC terms were ‘condensed chromosomes’ and ‘chromosomes, centromeric region’, which are known to be related with chromatin remodeling and genome stability (10-12). Exome sequencing has been used to identify a spontaneous mutation in MCM4 (MCM4d573h) in Spontaneous dominant leukemia (SDL) mice. As a part of the MCM2–7 heterohexameric complex, MCM4 plays an important role in gaining access to the DNA origin prior to the S phase, and is also the core replication helicase, which unwinds DNA at the replication fork. A study on Saccharomyces cerevisiae showed that the MCM4d573h mutation produced a type of bioactive helicase. These data support a model in which the chromosomal abnormalities of the SDL mice are due to the binding of MCM4D573H to the MCM complex, resulting in its inactivation. The dominant allele of MCMs is compatible with viability; however, it exerts significant carcinogenic effects and causes chromosomal abnormalities (10,43,44). Overall, the analysis of pathways and GO enrichment showed that MCM4 and its co-expressed genes played an important role in HCC carcinogenesis.
There are some limitations in the study. To elucidate the biological function of MCM4, further in vitro and in vivo investigations are required. We will continue to study the mechanism of MCM4 on HCC in the future work.
In this study, we systematically analyzed the expression, methylation status, mutations, CNAs, co-expression genes, and prognostic value of MCM4 in human hepatocarcinoma using online bioinformatics platforms and network tools. Multiomics analysis showed that MCM4 was upregulated and negatively correlated with the clinical prognosis of HCC. The overexpression of MCM4 may be regulated by promoter methylation and CNAs. Current findings revealed the importance of MCM4 expression and the associated pathways in cancer progression. Our results suggest that MCM4 may be used as a potential therapeutic target and prognostic biomarker for liver cancer.
Acknowledgments
We would like to thank Professor Lin Wo for offering expert advice concerning this study.
Funding: This work was supported by the Medical Scientific Research Foundation of the Guangdong Province of China (B2018064); the Shenzhen Sanming Project (SZSM201612041); the Shenzhen Science and Technology Innovation Commission Project (GJHZ20180420180754917, ZDSYS20190902092855097); and (JCYJ20170306170933370, JCYJ20180508152437368).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/jgo-20-574
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jgo-20-574
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jgo-20-574). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Written informed consent was obtained prior to study. This study was approved by the Ethic Committee of Affiliated Dongguan People’s Hospital, Southern Medical University. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Global Burden of Disease Cancer Collaboration, Fitzmaurice C, Akinyemiju TF, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2016: A systematic analysis for the Global Burden of Disease Study. JAMA Oncol 2018;4:1553-68. [Crossref] [PubMed]
- Ubhi T, Brown GW. Exploiting DNA Replication Stress for Cancer Treatment. Cancer Res 2019;79:1730-9. [Crossref] [PubMed]
- Macheret M, Halazonetis TD. DNA replication stress as a hallmark of cancer. Annu Rev Pathol 2015;10:425-48. [Crossref] [PubMed]
- Lin DI, Aggarwal P, Diehl JA. Phosphorylation of MCM3 on Ser-112 regulates its incorporation into the MCM2-7 complex. Proc Natl Acad Sci U S A 2008;105:8079-84. [Crossref] [PubMed]
- Vijayraghavan S, Schwacha A. The eukaryotic Mcm2-7 replicative helicase. Subcell Biochem 2012;62:113-34. [Crossref] [PubMed]
- Amaro Filho SM, Nuovo GJ, Cunha CB, et al. Correlation of MCM2 detection with stage and virology of cervical cancer. Int J Biol Markers 2014;29:e363-71. [Crossref] [PubMed]
- Huang B, Hu B, Su M, et al. Potential role of minichromosome maintenance protein 2 as a screening biomarker in esophageal cancer high-risk population in China. Hum Pathol 2011;42:808-16. [Crossref] [PubMed]
- Davies RJ, Freeman A, Morris LS, et al. Analysis of minichromosome maintenance proteins as a novel method for detection of colorectal cancer in stool. Lancet 2002;359:1917-9. [Crossref] [PubMed]
- Valverde LF, de Freitas RD, Pereira TA, et al. MCM3: A novel proliferation marker in oral squamous cell carcinoma. Appl Immunohistochem Mol Morphol 2018;26:120-5. [Crossref] [PubMed]
- Ishimi Y, Irie D. G364R mutation of MCM4 detected in human skin cancer cells affects DNA helicase activity of MCM4/6/7 complex. J Biochem 2015;157:561-9. [Crossref] [PubMed]
- Eissa S, Matboli M, Shehata HH, et al. MicroRNA-10b and minichromosome maintenance complex component 5 gene as prognostic biomarkers in breast cancer. Tumour Biol 2015;36:4487-94. [Crossref] [PubMed]
- Cai HQ, Cheng ZJ, Zhang HP, et al. Overexpression of MCM6 predicts poor survival in patients with glioma. Hum Pathol 2018;78:182-7. [Crossref] [PubMed]
- Ban X, Yan J, Yu S, et al. High minichromosome maintenance protein 7 proliferation indices: A powerful predictor of progression in pancreatic neuroendocrine neoplasms without distant metastasis at the time of surgery. Hum Pathol 2019;85:101-11. [Crossref] [PubMed]
- Yang JY, Li D, Zhang Y, et al. The expression of MCM7 is a useful biomarker in the early diagnostic of gastric cancer. Pathol Oncol Res 2018;24:367-72. [Crossref] [PubMed]
- Quaglia A, McStay M, Stoeber K, et al. Novel markers of cell kinetics to evaluate progression from cirrhosis to hepatocellular carcinoma. Liver Int 2006;26:424-32. [Crossref] [PubMed]
- Sun M, Wu G, Li Y, et al. Expression profile reveals novel prognostic biomarkers in hepatocellular carcinoma. Front Biosci (Elite Ed) 2010;2:829-40. [PubMed]
- Yang Q, Xie B, Tang H, et al. Minichromosome maintenance 3 promotes hepatocellular carcinoma radioresistance by activating the NF-κB pathway. J Exp Clin Cancer Res 2019;38:263. [Crossref] [PubMed]
- Xiang XH, Yang L, Zhang X, et al. Seven-senescence-associated gene signature predicts overall survival for Asian patients with hepatocellular carcinoma. World J Gastroenterol 2019;25:1715-28. [Crossref] [PubMed]
- Liu M, Hu Q, Tu M, et al. MCM6 promotes metastasis of hepatocellular carcinoma via MEK/ERK pathway and serves as a novel serum biomarker for early recurrence. J Exp Clin Cancer Res 2018;37:10. [Crossref] [PubMed]
- Qu K, Wang Z, Fan H, et al. MCM7 promotes cancer progression through cyclin D1-dependent signaling and serves as a prognostic marker for patients with hepatocellular carcinoma. Cell Death Dis 2017;8:e2603. [Crossref] [PubMed]
- Zhou YM, Zhang XF, Cao L, et al. MCM7 expression predicts post-operative prognosis for hepatocellular carcinoma. Liver Int 2012;32:1505-9. [Crossref] [PubMed]
- Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia 2004;6:1-6. [Crossref] [PubMed]
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007;9:166-80. [Crossref] [PubMed]
- Tang Z, Kang B, Li C, et al. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019;47:W556-60. [Crossref] [PubMed]
- Shin G, Kang TW, Yang S, et al. GENT: Gene Expression database of Normal and Tumor tissues. Cancer Inform 2011;10:149-57. [Crossref] [PubMed]
- Russi S, Calice G, Ruggieri V, et al. Gastric normal adjacent mucosa versus healthy and cancer tissues: Distinctive transcriptomic profiles and biological features. Cancers 2019;11:1248. [Crossref] [PubMed]
- Lian Q, Wang S, Zhang G, et al. HCCDB: A database of hepatocellular carcinoma expression atlas. Genomics Proteomics Bioinformatics 2018;16:269-75. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207-10. [Crossref] [PubMed]
- Goldman M, Craft B, Hastie M, et al. The UCHCC Xena platform for public and private cancer genomics data visualization and interpretation. bioRxiv 326470 (Preprint). 2019 (cited 2020 Oct 29). Available online: https://doi.org/ [Crossref]
- Goldman M, Craft B, Zhu J, et al. The UCHCC Xena system for cancer genomics data visualization and interpretation. Bioinform Syst Biol 2017;77:abstract 2584.
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Szász AM, Lánczky A, Nagy Á, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget 2016;7:49322-33. [Crossref] [PubMed]
- Zhou G, Soufan O, Ewald J, et al. NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res 2019;47:W234-41. [Crossref] [PubMed]
- Vogel A, Saborowski A. Current strategies for the treatment of intermediate and advanced hepatocellular carcinoma. Cancer Treat Rev 2020;82:101946. [Crossref] [PubMed]
- Huang XP, Rong TH, Wu QL, et al. MCM4 expression in esophageal cancer from southern China and its clinical significance. J Cancer Res Clin Oncol 2005;131:677-82. [Crossref] [PubMed]
- Winnepenninckx V, Lazar V, Michiels S, et al. Gene expression profiling of primary cutaneous melanoma and clinical outcome. J Natl Cancer Inst 2006;98:472-82. [Crossref] [PubMed]
- Kikuchi J, Kinoshita I, Shimizu Y, et al. Minichromosome maintenance (MCM) protein 4 as a marker for proliferation and its clinical and clinicopathological significance in non-small cell lung cancer. Lung Cancer 2011;72:229-37. [Crossref] [PubMed]
- Sanada H, Seki N, Mizuno K, et al. Involvement of dual strands of miR-143 (miR-143-5p and miR-143-3p) and their target oncogenes in the molecular pathogenesis of lung adenocarcinoma. Int J Mol Sci 2019;20:4482. [Crossref] [PubMed]
- Li C, Liu X, Liu Y, et al. Keratin 80 promotes migration and invasion of colorectal carcinoma by interacting with PRKDC via activating the AKT pathway. Cell Death Dis 2018;9:1009. [Crossref] [PubMed]
- Zhai DK, Liu B, Bai XF, et al. Identification of biomarkers and pathway-related modules involved in ovarian cancer based on topological centralities. J BUON 2016;21:208-20.
- Bagley BN, Keane TM, Maklakova VI, et al. A dominantly acting murine allele of MCM4 causes chromosomal abnormalities and promotes tumorigenesis. PLoS Genet 2012;8:e1003034. [Crossref] [PubMed]
- Shima N, Alcaraz A, Liachko I, et al. A viable allele of MCM4 causes chromosome instability and mammary adenocarcinomas in mice. Nat Genet 2007;39:93-8. [Crossref] [PubMed]
(English Language Editor: J. Jones)


