
ACTR3 promotes cell migration and invasion by inducing epithelial mesenchymal transition in pancreatic ductal adenocarcinoma
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is one of the most fatal malignancies, and is the fourth and sixth leading cause of cancer-related death in the USA and in China, respectively (1-3). Surgery is the only effective treatment of PDAC; however, approximately 20% of patients are offered radical surgery upon diagnosis due to arteriovenous invasion or distant metastasis (4-6). The 5-year survival rate of PDAC patients is <8% (7). Postoperative recurrence and metastasis are the major causes affecting the clinical prognosis of PDAC patients. The median survival of PDAC patients who have undergone surgical resection is less than 20 months, and approximately 20% of all resected PDAC patients survive >5 years (8-10).
The underlying mechanism of recurrence and metastasis of PDAC is currently poorly understood. Therefore, it is important to elucidate the molecular mechanisms responsible for the pathogenesis of recurrence and metastasis to optimize therapeutic strategies and improve the clinical prognosis for patients with PDAC.
Cancer metastasis is a multistage process, and includes invasion into surrounding tissue, transit into the blood or lymph system, and growth at a distant new site (11). Increased tumor cell motility is required during the process (12), which is driven by cycles of actin polymerization. Evidence has indicated that the polymerization of actin into filaments drives cell movement (13). Arp2/3 (actin-related proteins 2 and 3) is a multimeric complex of actin-binding proteins that play an essential role in the polymerization of actin into filaments, which is necessary not only for cell mobility but also for the maintenance of cell morphology (14).
Arp3, also known as actin-related proteins 3 (ACTR3), is a vital component of the Arp2/3 complex (15), which functions as an Actin-regulatory protein. Previously published studies have indicated that ACTR3 expression is increased in gastric cancer, squamous cell carcinoma, and colorectal cancer, and promotes tumor development (16-19). In addition, ACTR3 can combine with profilin-1 to regulate the formation of lamellipodia and filopodia with the assistance of LIM domain only 2 (LMO2) in breast cancer (20). In the present study, we found that ACTR3 was upregulated in human PDAC tissue by mRNA microarray analysis. However, to date, the role and the molecular mechanism of ACTR3 in PDAC remain elusive.
In the present study, we analyzed ACTR3 expression in PDAC tissue and the relationship of ACTR3 expression with clinical prognosis using bioinformatics. In vitro, the role of ACTR3 in the development of PDAC was examined. Our study may help to elucidate the specific mechanisms underlying the metastasis of PDAC.
We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/jgo-21-609).
Methods
mRNA microarray
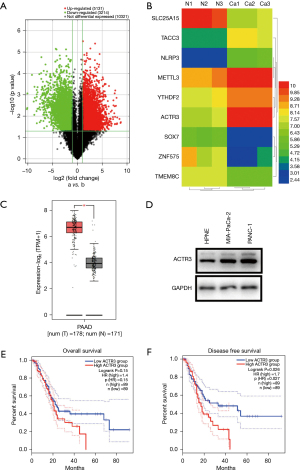
Three pairs of PDAC tissues and non-cancerous pancreatic tissues were selected for the mRNA microarray analysis. Total RNA was extracted and amplified and labeled with cyanine 3-CTP after the quality was assessed. TheCy3-CTP-labeled target was hybridized to microarrays (Kanchen Bio-tech, Shanghai, China) for 14 h. The microarrays were then scanned and raw signal intensities were collected. Differential RNAs were identified and plotted as a volcano map and heatmap (Figure 1A,1B).
All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional ethics board of the Affiliated Hospital of Jiangnan University (No. LS2019060), and informed consent was taken from all patients.
Bioinformatics
The Gene Expression Profiling Interactive Analysis (GEPIA2) database (http://gepia2.cancer-pku.cn/#index) was used to analyze the differences in ACTR3 expression between PDAC and adjacent non-cancerous tissue, and the relationship of ACTR3 expression with clinical prognosis.
Cell culture
PDAC cell lines (MIA-PaCa-2 and PANC-1) and the normal human pancreatic ductal cell line (HPNE) were purchased from Shanghai Cell Bank (Shanghai, China), and were cultured in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin/streptomycin (Gibco) in a humidified incubator with 5% CO2 at 37 °C.
Transient transfection of siRNAs
ACTR3-knockdown small-interfering RNA (siRNA) was synthesized by ProteinBio (Nanjing, China). The sequences of ACTR3siRNAs and the negative control (NC) are shown in Table 1. Transient transfections were performed using Lipofectamine 3000 (Life Technologies) according to the manufacturer’s instructions. ACTR3 expression was examined by Western blotting.
Table 1
| siRNAs | Sense (5'-3') | Antisense (5'-3') |
|---|---|---|
| ACTR3 si-1 | CCCUUCCUGUAUUGCUAUUAATT | UUAAUAGCAAUACAGGAAGGGTT |
| ACTR3 si-2 | CGUCCUCUCUACAAGAAUAUUTT | AAUAUUCUUGUAGAGAGGACGTT |
| ACTR3 si-3 | CCUCCAUUGAAUACUCCAGAATT | UUCUGGAGUAUUCAAUGGAGGTT |
| Negative control | UUCUCCGAACGUGUCACGUdTdT | ACGUGACACGUUCGGAGAAdTdT |
ACTR3, actin-related protein 3; siRNA, small-interfering RNA.
Phalloidin staining
Cells seeded in discs were fixed with 4% paraformaldehyde for 30 min. The cells were then incubated with 0.5% Triton X-100 for 10 min. After washing with phosphate-buffered saline, cells were stained with YF488-labeled phalloidin (US Everbright) for 30 min and 4',6-diamidino-2-phenylindole for 10 min, and then examined under a confocal microscope (Nikon, New York, USA).
Cell Counting Kit-8 (CCK-8) assay
Cells (2×103/well, 100 µL) were inoculated with complete medium in 96-wellplates. A total of 10 µL CCK-8 reagent (Dojindo, Tokyo, Japan) was added to each well at 24, 48, 72, and 96 h, respectively, and incubated for 1 h at 37 °C. Absorbance at 450 nm was calculated using a spectrophotometer.
Transwell assay
For the migratory capacity evaluation, 4×104cells were seeded into the upper chamber, and suspended and incubated in serum-free medium. A total of 800 µL complete medium containing 10% FBS was added to the bottom chamber. After 24 h of incubation, the chamber was fixed with 4% paraformaldehyde for 30 min. The upper cells were wiped away with cotton swabs and then stained with a 0.1% crystal violet solution for 20 min. Cells in 3–5 random fields were counted under ×100 magnification. All experiments were repeated 3 times.
For the invasive capacity evaluation, Matrigel (BD) was added to the upper chamber at a dilution of 1:8, and incubated at 37 °C for 1 h; 4×104 cells were seeded into the upper chamber, and suspended and incubated in serum-free medium. A total of 800 µL complete medium containing 10% FBS was added to the bottom chamber. After 24 h of incubation, the chamber was fixed with 4% paraformaldehyde for 30 min. The upper cells were wiped away with cotton swabs and then stained with a 1% crystal violet solution for 20 min. Cells in 3–5 random fields were counted under ×100 magnification. All experiments were repeated 3 times.
Western blotting
Cells were collected and proteins were extracted with radioimmunoprecipitation assay lysis buffer. Proteins were subjected to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane (Millipore). The membranes were then incubated with primary antibodies overnight at 4 °C after blocking with 5% non-fat milk for 2 h at room temperature. The primary antibodies were as follows: ACTR3 (1:2,000; ABclonal), GAPDH (1:20,000; ABclonal), vimentin (1:1,000; CST), E-cadherin (1:1,000; CST), N-cadherin (1:1,000; CST), and Snail (1:1,000; CST). After washing with (phosphate buffered saline-tween 20) PBST3 times, membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (1:5,000; Santa Cruz Biotechnology) for 2 h at room temperature. After washing with PBST 3 times, membranes were developed using an enhanced chemiluminescence detection system (UE Biotech, Suzhou, China).
Statistical analysis
The statistical analysis was performed using GraphPad Prism (version 6.01). Quantitative data are presented as the mean ± standard deviation, and differences between groups were analyzed using Student’s t-tests. P<0.05 was considered statistically significant.
Results
Upregulation of ACTR3 in PDAC and its association with poor prognosis
Differentially expressed RNAs were examined in 3 pairs of PDAC tissues and non-cancerous pancreatic tissues using high-throughput RNA sequencing. In total, 8,345 different genes were identified, with a statistical significance threshold at a fold change (FC) ≥2 and a false discovery rate (FDR) <0.05. Among these genes, there were 5,131 u-regulated RNAs and 3,214 downregulated RNAs, and ACTR3expression was 7-fold higher in PDAC tissues (FC =7.847, P=0.008, FDR =0.041) (Figure 1A,1B).
To assess the clinical significance of ACTR3, the GEPIA2 database was used for the analysis. mRNA levels of ACTR3 were analyzed in 179 PDAC tissues and 171 adjacent non-cancerous pancreatic tissues, and the results showed that ACTR3 was significantly upregulated in PDAC tissues (P<0.05) (Figure 1C). We further confirmed that ACTR3 was upregulated in PDAC cells compared with the HPNE cell line (Figure 1D). Survival analysis in 178 PDAC patients revealed that, although there was no relation between ACTR3 expression and overall survival (P=0.15) (Figure 1E), higher ACTR3 expression was predictive of shorter disease-free survival (DFS) (P=0.026) (Figure 1F). To data indicated that ACTR3 expression was upregulated in PDAC tissues and cell lines, and was closely associated with clinical prognosis.
No effect of ACTR3 on proliferation in PDAC cells
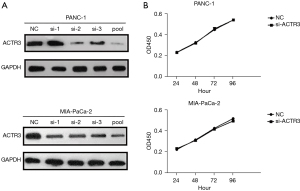
To elucidate the role of ACTR3on PDAC development, PANC-1 and MIA-PaCa-2 cells were transfected with ACTR3 siRNAs or an NC. The results indicated that individual siRNA showed different efficacy in the knockdown of ACTR3 expression compared with the NC, and pooled siRNAs showed the highest inhibitory efficacy of ACTR3 expression (Figure 2A). Therefore, pooled siRNAs were used for the following study. CCK-8 assay indicated no significant differences in the proliferation ability in PANC-1 cells and MIA-PaCa-2 cells transfected with ACTR3 siRNA and NC cells (Figure 2B).
ACTR3 promotes the migration and invasion of PDAC cells in vitro
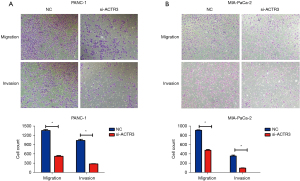
Transwell migration and invasion assays were conducted to determine the role of ACTR3 on cell migration and invasion. As shown in Figure 3A,3B, ACTR3 knockdown significantly repressed the migration of PANC-1 and MIA-PaCa-2 cells compared with the NC cells, and fewer ACTR3 siRNA-transfected PANC-1 and MIA-PaCa-2 cells migrated to the lower side of the membrane. Similarly, the knockdown of ACTR3 significantly inhibited the invasion of PANC-1 and MIA-PaCa-2 cells, and fewer ACTR3 siRNA-transfected PANC-1 and MIA-PaCa-2 cells invaded to the lower side of the membrane (Figure 3A,3B). The data indicated that ACTR3 promoted the migration and invasion of PANC-1 and MIA-PaCa-2 cells.
ACTR3 causes F-actin redistribution and promotes epithelial mesenchymal transition (EMT) in PDAC cells
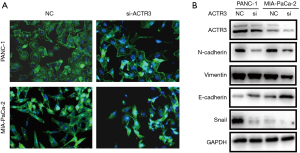
Actin is involved in many important cellular functions, including cell movement, cell division, and cytokinesis. In addition, EMT process is closely associated with cell migration and invasion. Therefore, we examined F-actin distribution and EMT-related proteins by phalloidin staining and Western blotting, respectively. As shown in Figure 4A, the knockdown of ACTR3 resulted in morphological changes and F-actin redistribution. Moreover, after ACTR3 knockdown, the expressions of N-cadherin and vimentin were downregulated, while the expression of E-cadherin was upregulated. Additionally, Snail was decreased when ACTR3 expression was downregulated (Figure 4B). These results suggest that ACTR3 promotes PDAC cell metastasis by regulating F-actin redistribution and the EMT process.
Discussion
Metastasis leads to approximately 90% of cancer-related deaths, and is considered one of the major obstacles in the treatment of cancers (21). The aggressive cell motility and deformability is the result of disassembly of actin filaments, together with actin polymerization (22,23). ACTR3 is an important member of the actin-related protein complex, which is involved in cell mobility and the EMT process. ACTR3 expression has been reported to be increased in a variety of tumors. However, the expression levels and the role of ACTR3 in the development of PDAC have not been reported.
In the current study, we examined the expression levels of ACTR3 in PDAC tissues and adjacent non-cancerous tissues. Our findings indicate that ACTR3 expression was significantly upregulated in PADC tissues Consistent with this result, the expression of ACTR3 in PDAC cell lines was significantly higher than that in HPNE cells. Survival analysis in 178 PDAC patients based on the GEPIA2 database revealed that increased ACTR3 expression was predictive of shorter DFS. Taken together, these findings indicate that ACTR3 has the potential to be a therapeutic target for PDAC.
To elucidate the exact role of ACTR3 in PDAC development, we observed the behavioral changes of PDAC cells by knocking down the expression of ACTR3. The results indicated that the invasive and migratory capacities of PANC-1 and MIA-PaCa-2 cells were significantly repressed uponACTR3 knockdown. However, ACTR3 knockdown had no effect on the proliferation of PDAC cells. Based on these results, we speculated that ACTR3 was associated with PDAC metastasis.
Cancer metastasis is a multiple-stage process regulated by several signaling molecules (24). During the EMT process, epithelial cells acquire the migratory capacity of mesenchymal cells, which plays a crucial role during the early stage of cancer metastasis (25,26). Therefore, we speculate that ACTR3 can promote EMT process to advance the metastasis of PDAC. In the current study, we found that the expression of E-cadherin (an epithelial marker) was significantly (27,28) increased upon the knockdown of ACTR3 expression. In contrast, the expressions of N-cadherin and vimentin (mesenchymal markers) (27,28) were downregulated. The expression of Snail, a transcriptional repressor of E-cadherin (29,30), also decreased as ACTR3 expression was downregulated. We also found that the knockdown of ACTR3 caused the redistribution of F-actin and morphological changes in PDAC cells. Therefore, ACTR3 can promote the metastasis of PDAC through EMT. Based the findings above, we speculated that ACTR3 may promote the polymerization of actin into filaments, thus regulating cell morphology and mobility and eliciting PDAC metastasis cell.
The present study has some limitations. First, only an in vitro study was performed to examine the role of ACTR3 in PDAC. The role of ACTR3 should be verified in animal models in future studies. Second, the overexpression of ACTR3 in PDAC cells confirmed the promoting role of ACTR3 in PDAC metastasis. Finally, an in-depth mechanism study is needed to further identify therapeutic targets against PDAC metastasis.
In conclusion, our study demonstrated, for the first time, thatACTR3 was upregulated in PDAC, and was a metastasis-associated gene in PDAC. Additionally, ACTR3 expression was predictive of poor prognosis for PDAC patients. Therefore, ACTR3 may be a potential therapeutic target for PDAC metastasis.
Acknowledgments
Funding: This work was supported by Jiangsu Natural Science Foundation Project (No. BK20191142), Science and Technology Development Plan Project of Wuxi (No. N20202011), High-level Talent Training Project of Wuxi Taihu Talent Plan (No. BJ2020055), Scientific Research Project of Jiangsu Provincial Health Commission (No. Z2018022), Major Scientific Research Project of Wuxi Municipal Health Commission (No. Z201805), Research Team Construction Funding Project of Jiangnan University School of Medicine (No. 1286010242190110), Translational Medicine Research Project of Wuxi Municipal Health Commission (No. ZM002), Special Funding of Science and Education Enhancing Health of Wuxi (No. QNRC015), and Young Researcher Grants Program of Wuxi Health and Family Planning Commission (No. Q201621).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/jgo-21-609
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jgo-21-609
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jgo-21-609). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional ethics board of the Affiliated Hospital of Jiangnan University (No. LS2019060), and informed consent was taken from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Strobel O, Neoptolemos J, Jäger D, et al. Optimizing the outcomes of pancreatic cancer surgery. Nat Rev Clin Oncol 2019;16:11-26. [Crossref] [PubMed]
- Gilbert JW, Wolpin B, Clancy T, et al. Borderline resectable pancreatic cancer: conceptual evolution and current approach to image-based classification. Ann Oncol 2017;28:2067-76. [Crossref] [PubMed]
- Kenkel JA, Tseng WW, Davidson MG, et al. An Immunosuppressive Dendritic Cell Subset Accumulates at Secondary Sites and Promotes Metastasis in Pancreatic Cancer. Cancer Res 2017;77:4158-70. [Crossref] [PubMed]
- Kamisawa T, Wood LD, Itoi T, et al. Pancreatic cancer. Lancet 2016;388:73-85. [Crossref] [PubMed]
- Halbrook CJ, Lyssiotis CA. Employing Metabolism to Improve the Diagnosis and Treatment of Pancreatic Cancer. Cancer Cell 2017;31:5-19. [Crossref] [PubMed]
- Dal Molin M, Zhang M, de Wilde RF, et al. Very Long-term Survival Following Resection for Pancreatic Cancer Is Not Explained by Commonly Mutated Genes: Results of Whole-Exome Sequencing Analysis. Clin Cancer Res 2015;21:1944-50. [Crossref] [PubMed]
- Grasso C, Jansen G, Giovannetti E. Drug resistance in pancreatic cancer: Impact of altered energy metabolism. Crit Rev Oncol Hematol 2017;114:139-52. [Crossref] [PubMed]
- Griffin JF, Poruk KE, Wolfgang CL. Pancreatic cancer surgery: past, present, and future. Chin J Cancer Res 2015;27:332-48. [PubMed]
- Machesky LM. Lamellipodia and filopodia in metastasis and invasion. FEBS Lett 2008;582:2102-11. [Crossref] [PubMed]
- Yilmaz M, Christofori G. Mechanisms of motility in metastasizing cells. Mol Cancer Res 2010;8:629-42. [Crossref] [PubMed]
- Olson MF, Sahai E. The actin cytoskeleton in cancer cell motility. Clin Exp Metastasis 2009;26:273-87. [Crossref] [PubMed]
- Vartiainen MK, Machesky LM. The WASP-Arp2/3 pathway: genetic insights. Curr Opin Cell Biol 2004;16:174-81. [Crossref] [PubMed]
- Gross SR. Actin binding proteins: their ups and downs in metastatic life. Cell Adh Migr 2013;7:199-213. [Crossref] [PubMed]
- Zheng HC, Zheng YS, Li XH, et al. Arp2/3 overexpression contributed to pathogenesis, growth and invasion of gastric carcinoma. Anticancer Res 2008;28:2225-32. [PubMed]
- Yang ZL, Miao X, Xiong L, et al. CFL1 and Arp3 are biomarkers for metastasis and poor prognosis of squamous cell/adenosquamous carcinomas and adenocarcinomas of gallbladder. Cancer Invest 2013;31:132-9. [Crossref] [PubMed]
- Otsubo T, Iwaya K, Mukai Y, et al. Involvement of Arp2/3 complex in the process of colorectal carcinogenesis. Mod Pathol 2004;17:461-7. [Crossref] [PubMed]
- Yin X, Zhang Y, Guo S, et al. Large scale systematic proteomic quantification from non-metastatic to metastatic colorectal cancer. Sci Rep 2015;5:12120. [Crossref] [PubMed]
- Liu Y, Wu C, Zhu T, et al. LMO2 Enhances Lamellipodia/Filopodia Formation in Basal-Type Breast Cancer Cells by Mediating ARP3-Profilin1 Interaction. Med Sci Monit 2017;23:695-703. [Crossref] [PubMed]
- Gupta GP, Massagué J. Cancer metastasis: building a framework. Cell 2006;127:679-95. [Crossref] [PubMed]
- Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct 2007;36:451-77. [Crossref] [PubMed]
- Yamaguchi H, Lorenz M, Kempiak S, et al. Molecular mechanisms of invadopodium formation: the role of the N-WASP-Arp2/3 complex pathway and cofilin. J Cell Biol 2005;168:441-52. [Crossref] [PubMed]
- Yamazaki D, Kurisu S, Takenawa T. Regulation of cancer cell motility through actin reorganization. Cancer Sci 2005;96:379-86. [Crossref] [PubMed]
- van Zijl F, Zulehner G, Petz M, et al. Epithelial-mesenchymal transition in hepatocellular carcinoma. Future Oncol 2009;5:1169-79. [Crossref] [PubMed]
- Giannelli G, Koudelkova P, Dituri F, et al. Role of epithelial to mesenchymal transition in hepatocellular carcinoma. J Hepatol 2016;65:798-808. [Crossref] [PubMed]
- Hajra KM, Fearon ER. Cadherin and catenin alterations in human cancer. Genes Chromosomes Cancer 2002;34:255-68. [Crossref] [PubMed]
- Sommariva M, Gagliano N. E-Cadherin in Pancreatic Ductal Adenocarcinoma: A Multifaceted Actor during EMT. Cells 2020;9:1040. [Crossref] [PubMed]
- Batlle E, Sancho E, Francí C, et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol 2000;2:84-9. [Crossref] [PubMed]
- Cano A, Pérez-Moreno MA, Rodrigo I, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol 2000;2:76-83. [Crossref] [PubMed]
(English Language Editor: R. Scott)


