
Complement C5 is a novel biomarker for liver metastasis of colorectal cancer
Introduction
Colorectal cancer (CRC) is a notorious malignancy associated with a high incidence and poor prognosis (1). In 2020, there were more than 1.9 million colon cancer cases and 0.9 million deaths worldwide, and it is expected to affect 30 million people by 2040 (2). The liver is the most common site of CRC metastasis, and 15–25% of CRC patients have synchronous liver metastasis at the initial diagnosis (3). Regrettably, 15–25% of CRC patients develop heterochronous liver metastases following primary tumor resection, and 80–90% of these patients lose the opportunity for reoperation (4). According to previous data, there is only a 6.9-month median survival rate for patients with unresectable liver metastases (5,6), and a 30–57% 5-year survival rate for patients with resectable liver metastases (7). Thus, there is an urgent need to study the mechanisms of molecular biomarkers to predict liver metastases of colorectal cancer (LMCRC).
The microarray technique has been extensively applied in medicine and the life sciences. Bioinformatics of gene expression microarray data is an excellent tool to explore various molecular mechanisms, and hub genes from microarray can provide valuable hints for further fundamental research (8). Through high-throughput bioinformatics technology, some authors have uncovered that HOXD10, SLC13A2, OSM, and MMP3 may have potential as biomarkers for liver metastases associated with CRC (9-11). Other articles suggested that APOA1, APOB, APOE, etc. might be related to CRC metastasis (12-14). However, a shortage of clinical samples has inhibited further verification of hub gene availability in relevant research, and the deficiency of in vitro experiments could not uncover the relationship between the potential biomarkers and the progression of CRC.
In this study, we systematically analyzed three Gene Expression Series (GESs) containing CRC tissues and LMCRC using bioinformatics software and found that the complement and coagulation cascade signaling pathways were significantly enriched and the complement 5 (C5) might promote the progression of LMCRC, which is consistent with previous study (15). The distinction is that we not only conducted many further in-vitro and in-vivo investigations regarding C5 and LMCRC, but also independently verified the findings using matched tissues from LMCRC patients. The final results suggested that C5 could be a potential biomarker for poor progress in liver metastatic CRC patients. We present the following article in accordance with the MDAR reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-829/rc).
Methods
Data download for microarrays
The high-throughput gene expression profiles of CRC and LMCRC tissues were extracted from the Gene Expression Omnibus (GEO) database. Three independent datasets (GSE41258, GSE49355, and GSE81558) were analyzed, which contained 186 CRC tissues and 47 liver metastases, 20 CRC tissues and 19 liver metastases, 23 CRC tissues and 19 liver metastases, respectively.
Identification of differentially expressed genes (DEGs)
The DEGs between CRC and liver metastases were assessed using the online GEO2R tool (https://www.ncbi.nlm.nih.gov/geo/geo2r/). Statistical significance was determined by P<0.05 and |log fold change (FC)| ≥1. The DEGs in the three datasets were then screened using Venn software (https://bioinformatics.psb.ugent.be/webtools/Venn/). Genes with logFC ≥1 were considered up-regulated, and those with logFC ≤−1 were considered down-regulated.
Functional and pathway enrichment analysis
The Database for Annotation, Visualization, and Integrated Discovery (DAVID) bioinformatics resources (https://david.ncifcrf.gov/home.jsp) were utilized for annotation, visualization, and integrated discovery, including the biological process (BP), molecular function (MF), cellular component (CC), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. The cut-off criteria were set at P<0.05.
Protein-protein interaction (PPI) construction and module analysis
An interactive protein-protein interaction (PPI) network was built via the Search Tool for the Retrieval of Interacting Genes (STRING) (https://cn.string-db.org/) database to uncover the relationships between the DEGs. An interaction score ≥0.4 was set as the cut-off criterion, and the PPI network was then visualized using Cytoscape (version 3.9.1) software (National Institute of General Medical Sciences, Maryland, USA). Based on the genes in the network, we searched for hub genes via the Molecular Complex Detection (MCODE) (score cut-off =0.2, degree cut-off =2, maximum depth =100, and k-score =2) method.
RNA expression and survival analysis of hub genes
The University of ALabama at Birmingham Cancer (UALCAN) (http://ualcan.path.uab.edu/index.html) database was used to analyze the hub gene expression levels of colon adenocarcinoma (COAD) between normal colon tissues and primary tumors (CRC tissues). Statistical significance was defined as P<0.05.
RNA extraction, reverse transcription-polymerase chain reaction (RT-PCR), and quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Trizol reagent (Invitrogen, Carlsbad, CA, USA) was used for the extraction of total RNA from cell lines. The complementary DNA and subsequent experiments were synthesized and performed using the Prime Script™ RT Reagent Kit (Takara Bio, Dalian, China). The primer sequences were as follows: glyceraldehyde 3-phosphate dehydrogenase (GAPDH) forward, 5'-ACCCACTCCTCCACCTTTG-3'; and reverse, 5'-CTGTAGCCAAATTCGTTGTCAT-3'; C5 forward, 5'-ACATTACGAGTGGTGCCAGA-3'; and reverse, 5'-TGGGGAGGTGGGTTAGGATA-3'.
Cell lines and cell transfection
The COAD cell lines, RKO, HT-29, HCT116, SW480, and SW620, were obtained from the National Collection of Authenticated Cell Cultures. The cells were cultured in a medium supplemented with 10% fetal bovine serum (FBS, Thermo Fisher Scientific, Waltham, MA, USA). The cells were maintained at 37 ℃ in a humidified incubator containing 5% CO2. According to the manufacturer’s instructions, a total of 1×104 cells/mL were plated in 96-well plates for 24 h and transfected with 2.5 nm short hairpin RNA/negative control (shRNA/NC) with lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA) when they reached 40–60% confluence in each well. After 6 hours of transfection, the medium was replaced with a fresh medium containing 10% FBS. After 24 hours, the cells were harvested for follow-up experiments. The shRNA sequences were as follows: Sh-h-C5: forward, 5'-CCGGGCCCGAGAGAACAGCTTATCTCGAGATAAGCTGTTCTCTCGGGCTTTTTG-3'; reverse, 5'-AATTCAAAAAGCCCGAGAGAACAGCTTATCTCGAGATAAGCTGTTCTCTCGGGC-3'.
Western blot
A radioimmunoprecipitation assay buffer (Beyotime Institute of Biotechnology, Haimen, China) was used for lysate preparation, and protein was measured with a bicinchoninic acid assay kit (Pierce; Thermo Fisher Scientific, Inc.). A total of 200-µg/well protein was electrophoresed in 10% sodium dodecyl sulfate (SDS) polyacrylamide gels and then transferred onto polyvinylidene fluoride (PVDF) membranes. The membranes were blocked with 5% fat-free milk at room temperature and incubated with a specific primary antibody at 4 ℃ overnight. Next, the membranes were incubated with secondary antibodies at room temperature for 2 h. Antibody binding was detected by an enhanced chemiluminescence western blotting substrate (Pierce; Thermo Fisher Scientific, Inc.). The primary antibodies used were as follows: C5 (dilution, 1:1,000; cat. No. ab275933; Abcam, Cambridge, UK) and GAPDH (dilution, 1:10,000; cat. no. AP0063; Bioworld Technology, Inc., St. Louis Park, MN, USA).
Cell proliferation, colony formation, and invasion assays
For the cell proliferation assay, cells (100 µL aliquots) were seeded in 96-well plates at a density of 2,000 cells/well, and 10 µL cell counting kit-8 (CCK-8) solution (Dojindo, Kumamoto, Japan) was added at the indicated time points. Cell viability was determined by measuring the absorbance of 450 nm at the indicated time points of 2, 24, 48, and 72 h, respectively.
For the colony formation assay, after knockdown of C5 for 2 weeks, 500–1,000 SW480 or SW620 cells were plated on six-well plates and cultured at 37 ℃ for about 14 days. The cell colonies were then fixed with ice-cold methanol, stained with 0.5% crystal violet, and photographed. The number of clones was counted using Image-Pro Plus v6.2 (Media Cybernetics, Inc., Silver Spring MD, USA).
For the invasion assay, a 24-well Transwell chamber with Matrigel (BD Biosciences) was prepared. Firstly, 1×105 cells/w were added into the upper chamber with 100 µL Dulbecco’s modified Eagle medium (DMEM) supplemented with 1% FBS and then incubated in a serum-free culture medium for 48 h. Next, we removed the Matrigel and the non-migrating cells in the upper chamber. The cells on the bottom were fixed with 4% paraformaldehyde. Subsequently, the cells were stained with Giemsa and counted from five microscopic (×200) fields.
Wound‑healing assay
The cell migration of SW480 and SW620 was assessed by a wound-healing assay. After knockdown of C5 for 2 weeks, cells that were in the logarithmic phase were digested. The cells were then plated on 12-well plates at a density of 4×105 cells by using sterile tips, wounds were created and the migration distances were recorded by an inverted microscope (Olympus, Tokyo, Japan) at 0, 24, and 48 hours.
Tumor xenograft mouse model
Animal experiments were approved by the Institutional Animal Care and Use Committee at Fudan University (No. 2021753), in compliance with Chinese National Standard (GBT35823-2018) for the care and use of animals. Male C57/Bl6 mice (6 weeks, 20±2 g) were selected and purchased from SLAC (Shanghai, China). Stable knockdown of C5 in SW480 or SW620 cells was established by stably transfected with Sh2_C5. Then, 5×106 cells were injected subcutaneously into the dorsal right flank of mice (three mice per group). At the endpoint (2 weeks post injection), the mice were sacrificed and tumors were excised and weighed. Tumor volume was measured by vernier caliper.
Immunohistochemistry (IHC) assay and evaluation of the IHC staining
IHC staining was conducted based on a two-step protocol (Novolink Polymer Detection System, Novocastra, UK). After antigen retrieval, the slides were incubated with antibodies overnight at 4 ℃, followed by 30 minutes of incubation with a secondary antibody (GK500705, Gene Tech, Shanghai, China) at 37 ℃. A 3,3-diaminobenzidine solution and Mayer’s hematoxylin were used for incubating cells and counterstaining. The corresponding negative controls were included in all assays without primary antibodies. A consensus was reached after three independent pathologists assessed the IHC staining without knowledge of the clinical or pathologic features of the case. Three representative microscope fields were captured under high magnification (×200) using Leica QWin Plus v3 software (Leica Microsystems, Wetzlar, Germany). The settings for each photograph were identical.
Patients’ specimens
A total of 20 paired fresh LMCRC and CRC tissues (12 tissues were used for qRT-PCR analysis and the remaining eight tissues were used for IHC analysis) were collected from CRC patients who received homochronous colectomy and hepatic resection from 2016 to 2020 in Zhongshan Hospital of Fudan University, China. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Review Board of Zhongshan Hospital of Fudan University (No. B2020-348R) and informed consent was taken from all the patients.
Statistical analysis
The results were presented as mean ± standard error of the mean (SEM). Statistical analyses were performed using SPSS 22.0 (SPSS Inc., IL, USA) and GraphPad 8.0 (La Jolla, CA, USA) software. All experiments were performed at least three times. Statistical significance was determined with asterisks and P values using the Student’s t-test (P<0.05, P<0.01, and P<0.001 were marked by *, **, and ***, respectively).
Results
Identification of aberrantly expressed genes
In this study, we downloaded three GEO datasets and screened DEGs via the online GEO2R tool. The three microarrays contained a total of 229 CRC tissues and 85 liver metastases. The findings revealed that 100, 170, and 215 genes were up-regulated whereas 117, 166, and 92 genes were down-regulated in GSE41258, GSE49355, and GSE81558, respectively (Figure 1A). According to the online Venn diagram, there were 92 common DEGs in the three microarrays, including 70 up-regulated genes and 22 down-regulated genes (Figure 1B,1C; Table 1; available at https://cdn.amegroups.cn/static/public/jgo-22-829-1.xlsx).
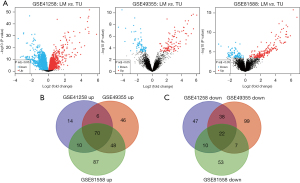
Table 1
| Regulation | DEGs |
|---|---|
| Up-regulated (n=70) | FGA, ORM1, IGFBP1, AMBP, CPS1, LECT2, HGD, SLC2A2, CFHR2, ITIH3, RBP4, APCS, FGG, C4BPA, ALB, PLG, AOX1, UGT2B15, ALDOB, CP, SERPINC1, SERPIND1, ALDH8A1, CRP, CYP2C8, SPP1, ITIH4, AADAC, F2, APOA1, HPR, GC, SERPINA1, SERPINA10, APOA2, CYP3A4, HPD, APOC1, LBP, ASGR1, COLEC11, AHSG, ADH1A, ARG1, FGL1, HPX, FMO3, TF, C8A, HRG, C5, APOC3, F9, KNG1, SERPINA3, ADH1B, CPB2, MBL2, CYP2E1, AGXT, APOE, APOB, ITIH2, TTR, VNN1, APOH, CDH2, GATM, FGB, HP |
| Down-regulated (n=22) | FOXF1, MMP2, FCGBP, CXCL14, DKK3, PLN, MMP1, ACTG2, PDZD2, MYH11, FBLN1, CHL1, JCHAIN, TMEM158, GREM1, MMP3, VWF, CAV1, CTSK, SPINK4, DES, SPARCL1 |
DEGs, differentially expressed genes; GEO, Gene Expression Omnibus; CRC, colorectal cancer; LMCRC, liver metastasis of colorectal cancer.
Gene Ontology (GO) and KEGG enrichment analysis
We discovered 183 significant enrichment terms in the up-regulated group through the online DAVID software, including BP [117], MF [39], and CC [27] (available at https://cdn.amegroups.cn/static/public/jgo-22-829-2.xlsx). In terms of BP, it was discovered that DEGs were considerably abundant in cellular protein metabolism, posttranslational protein modification, platelet degranulation, negative regulation of endopeptidase activity, blood coagulation, and other processes. Regarding MF, the DEGs showed a marked increase in protein binding, identical protein binding, receptor binding, serine-type endopeptidase inhibitor activity, heparin binding, and other molecules. As for CC, extracellular areas, extracellular exosomes, extracellular space, blood microparticles, endoplasmic reticulum lumen, and other structures were significantly enriched in DEGs. The top 10 GO terms are shown in Figure 2A-2C.
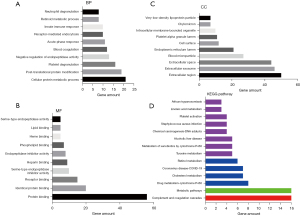
Also, KEGG pathway analysis revealed that DEGs were abundant in 14 pathways, including complement and coagulation cascades, metabolic pathways, cytochrome P450 drug metabolism, cholesterol metabolism, coronavirus disease (COVID-19), and retinol metabolism (Figure 2D; Table 2; Table S1). We also identified 30 significant enrichment terms in the down-regulated group, including BP [19], MF [6], and CC [5] (available at https://cdn.amegroups.cn/static/public/jgo-22-829-3.xlsx). As for the KEGG pathway, the down-regulated groups were enriched in two pathways, including Rheumatoid arthritis and COVID-19 (Figure S1; Table S2).
Table 2
| Category | Term | Count | P value | FDR |
|---|---|---|---|---|
| KEGG_PATHWAY | Complement and coagulation cascades | 16 | 9.12E-19 | 7.94E-17 |
| KEGG_PATHWAY | Metabolic pathways | 16 | 0.038935 | 0.25238 |
| KEGG_PATHWAY | Drug metabolism - cytochrome P450 | 8 | 2.38E-07 | 1.03E-05 |
| KEGG_PATHWAY | Cholesterol metabolism | 7 | 5.21E-07 | 1.51E-05 |
| KEGG_PATHWAY | Coronavirus disease - COVID-19 | 7 | 0.002845 | 0.035355 |
| KEGG_PATHWAY | Retinol metabolism | 6 | 5.64E-05 | 0.001132 |
| KEGG_PATHWAY | Tyrosine metabolism | 5 | 6.51E-05 | 0.001132 |
| KEGG_PATHWAY | Metabolism of xenobiotics by cytochrome P450 | 5 | 0.001303 | 0.01889 |
| KEGG_PATHWAY | Alcoholic liver disease | 5 | 0.011117 | 0.107463 |
| KEGG_PATHWAY | Chemical carcinogenesis - DNA adducts | 4 | 0.008678 | 0.094372 |
| KEGG_PATHWAY | Staphylococcus aureus infection | 4 | 0.02108 | 0.156711 |
| KEGG_PATHWAY | Platelet activation | 4 | 0.040613 | 0.25238 |
| KEGG_PATHWAY | Linoleic acid metabolism | 3 | 0.013595 | 0.118276 |
| KEGG_PATHWAY | African trypanosomiasis | 3 | 0.021615 | 0.156711 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; FDR, false discovery rate.
PPI network construction and module analysis
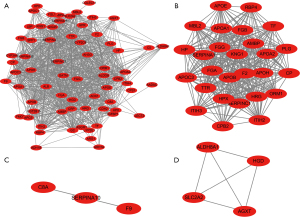
The STRING database was used to build the PPI network of up-regulated DEGs genes, which was then visualized using Cytoscape software (Figure 3A). MCODE was further used to analyze hub genes, including 27 nodes and 319 edges (Figure 3B; available at https://cdn.amegroups.cn/static/public/jgo-22-829-4.xlsx). The hub genes included ALDH8A1, SLC2A2, HGD, AGXT, F9, SERPINA10, and C8A (Figure 3C,3D). PPI revealed only four nodes and six edges for down-regulated DEGs genes, including MMP3, CTSK, MMP2, and MMP1 (Figure S2; available at https://cdn.amegroups.cn/static/public/jgo-22-829-5.xlsx).
C5 expression is increased in metastatic CRC
Although the above analysis did not classify C5 as a hub gene via MCODE, as an upstream gene of C8A in the complement system, it was also found to be related to other hub genes (Figure 3A,3C), and KEGG analysis in metastatic CRC revealed that the complement and coagulation cascades contributed most (Figure 2D); so, verification of C5 in The Cancer Genome Atlas (TCGA) was necessary. COAD samples demonstrated considerably higher C5 expression than normal samples, according to the UALCAN online data (Figure 4A). Additionally, C5 also showed increasing trends in more malignant cancer stages of COAD and was significantly correlated with the survival rates of patients (Figure 4B,4C; Table S3).
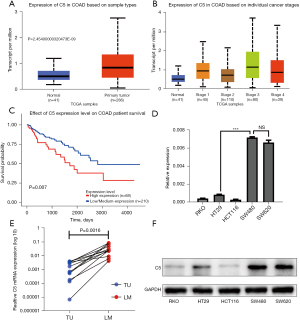
Real-time PCR indicated that the C5 mRNA levels in the SW620 and SW480 cell lines were significantly higher than those in the RKO, HT29, and HCT116 cell lines (Figure 4D). Meanwhile, 12 paired metastatic CRC samples in our hospital revealed that the mRNA levels of C5 were higher in liver metastasis tissues (Figure 4E). Western blot also revealed similar results in terms of the protein levels in the CRC cell lines (Figure 4F).
C5 promoted proliferation, migration, and invasion
Based on the C5 results from the above research, we knocked down C5 expression in SW480 and SW620 cells via lentivirus transfection (Figure 5A,5B); the CCK-8 kit revealed that the proliferation of SW480 and SW620 cells decreased significantly after knockdown of C5 compared to the control group in 2 days (Figure 5C,5D). Further, we performed a xenograft model assay through subcutaneously injecting the stable C5 knockdown and control cells into the flank of nude mice. The shC5 groups exhibited a much slower tumor growth rate than the control group. The volume and weight of tumors formed by C5-knockdown cells were also significantly decreased compared with the control group at the termination of the experiment (Figure 5E,5F). The wound-healing assay further proved the migration was notably reduced after 48 h (Figure 5G,5H). The transwell experiment also showed the invasion decreased markedly after knockdown of C5 was compared with the control group (Figure 6A,6B). Figure 6C,6D further demonstrated that the sh-C5 group’s colony formation ability declined significantly.

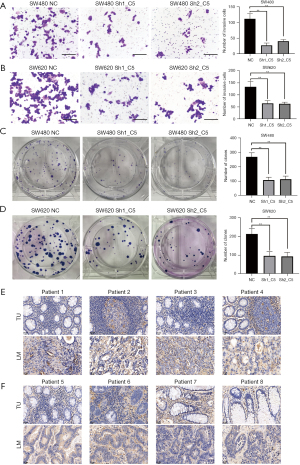
C5 protein expression was up-regulated in paired LMCRC tissues
Finally, we collected eight paired LMCRC tissues in our hospital to analyze their protein expression via an IHC assay. The results revealed higher C5 protein expression in LMCRC tissues (patients 1–6) compared to the paired primary CRC tissues (Figure 6E,6F). Also, two paired cases showed that C5 expression was similar between the primary CRC tissues and liver metastasis tissues (Figure 6F).
Discussion
Despite the continuous development of medical technology and the constantly updated guidelines, CRC is still the second-leading contributor to cancer-related deaths globally (1). Liver metastasis is a leading cause of death in patients with colorectal cancer. Around 20–25% of the patients present with LMCRC at the time of diagnosis, while up to 50% will develop LMCRC within three years after the diagnosis of the primary malignancy. Only around 20% of the LMCRC patients present amenable to resection with curative intent (16). When untreated, patients with LMCRC only have a median survival of 6 to 9 months (17). Complete tumor resection is the only treatment option with a potential for cure and long-term survival (16).
The occurrence and progression of CRC is the result of an interactive process of comprehensive factors. Combination of immunotherapy with other therapies, like antiangiogenic drugs has become a novel strategy to treat CRC (18). A good biomarker should predict not only prognosis but also the response to therapies. It has been established certain molecules are expressed at varying amounts in different stages of CRC (19), especially in the last stage, where the prognosis of LMCRC is poor (3,6). Therefore, it is essential to identify reliable metastatic biomarkers for CRC.
In the present study, we deliberately concentrated on the CRC microarray expression profile data. In our investigation, 85 liver metastasis tissues and 229 CRC tissues from three GSE datasets of the GEO database were included. A total of 92 DEGs, including 22 down-regulated genes and 70 up-regulated genes, were discovered. The GO and KEGG enrichment analysis using the online DAVID software showed that the metastatic process was a complex system with several function changes. The up-regulated DEGs were found to be particularly enriched in cellular protein metabolic process, posttranslational protein modification, platelet degranulation, negative regulation of endopeptidase activity, blood coagulation, acute-phase response, receptor-mediated endocytosis, innate immune response, retinoid metabolic process, neutrophil degranulation, and 114 other BPs. The up-regulated DEGs were also accumulated in 27 CCs, including the extracellular region, extracellular exosome, extracellular space, blood microparticle, endoplasmic reticulum lumen, cell surface, platelet alpha granule lumen, intracellular membrane-bounded organelle, chylomicron, very-low-density lipoprotein particle, etc. Additionally, the up-regulated DEGs were also found to be significantly enriched in 39 MF areas, including protein binding, identical protein binding, receptor binding, serine-type endopeptidase inhibitor activity, heparin binding, endopeptidase inhibitor activity, phospholipid binding, heme binding, lipid binding, serine-type endopeptidase activity, etc. KEGG pathway analysis showed that up-regulated DEGs were found in 14 aspects such as complement and coagulation cascades, metabolic pathways, drug metabolism-cytochrome P450, cholesterol metabolism, COVID-19, retinol metabolism, etc. Given the small amount of enrichment of down-regulated DEGs, the particular results were clarified in Figure S1 and Table S2.
Numerous interactions among the up-regulated DEGs were discovered by constructing PPI networks, and MCODE was then used to investigate hub genes such as ALDH8A1, SLC2A2, HGD, AGXT, F9, SERPINA10, and C8A (Figure 3C,3D). Our further investigation of the relationship between hub gene expression and the prognosis of COAD patients by UALCAN analysis yielded regrettable results. So, we shifted our minds to focus the results on GO, KEGG, and PPI; we found that complement and coagulation cascades were significantly enriched in the up-regulated DEGs, and C5 and C8A seemed to exert a key function in the PPI network construction (Figure 2D and Figure 3). Next, UALCAN showed that compared to the normal samples, C5 was considerably higher in the COAD samples (Figure 4A). Additionally, C5 was surprisingly associated with the survival of COAD patients (Figure 4C; Table S3).
The complement system is composed of more than 50 soluble proteins and membrane-bound proteins, including C1–C9, natural ingredients (C3aR, C5aR, CR2, etc.), complement receptors (CFI, CFH, etc.), and complement regulatory proteins (20,21). The complement system has been shown to play a complex role in various cancer types at different stages (22). On the one hand, the complement system plays a role in the direct killing effects on tumors via activation of the immune system; on the other hand, it promotes tumor progression, metastasis, immune escape, and immunosuppression, etc. through long-term low or high expression of complement-related factors in the tumor microenvironment system to maintain chronic local inflammation (23-25). The complement system can be activated through classical, lectin, or alternative pathways to further stimulate antigen-presenting cells (APC) to recognize tumor cells. Meanwhile, the formation of a membrane attack complex (MAC) by the complement system (namely complement-dependent cytotoxicity) can lead to tumor cell lysis (26,27). Therefore, future research will focus on complement-related gene expression changes in primary or metastatic tumors to search for new therapeutic strategies for malignant cancer.
One article reported that complement C5 or specifically C5AR1 deficiency completely inhibits the development of CRC tumorigenesis by recruiting myeloid-derived suppressor cells (MDSCs) to the inflamed colorectum and impairing CD8 (+) Treg (T) cells, and its mechanism revealed the close relationship between the loss of C5AR1 and chemokines [interleukin-6 (IL-6), IL-11, IL-27, etc.] (28). Another study reported that the C3AR and C5AR1-mediated signaling pathways promoted the transformation of the tumor microenvironment to tumor progression by activating the polarizing natural immune cells, inhibiting effector T cells, and releasing pro-tumor factors (29). C5AR1 has even been shown to inhibit T helper 1 (Th1) production and convert tumor-associated macrophages (TAMs) into M2 phenotype (23). Daugan et al. reported that increased expression of the C4-activating enzyme, C1s, by tumor cells is linked to a poor prognosis in clear cell renal cell carcinoma (CCRCC), and the primary mechanisms are a high infiltration of macrophages and T cells, complement cascade reaction, and the non-canonical method (30). Furthermore, one Nature communication article revealed that C3 could facilitate the lung metastasis of breast cancer through neutrophil recruitment and the synthesis of neutrophil extracellular traps (NETs), and preventing the Th2-Signal Transducer and Activator of Transcription 6 (STAT6)-C3-NETs cascade may mitigate breast cancer metastasis to the lungs (31).
Since previous articles have revealed that the complement system can play a key role in tumorigenesis and development, we shifted our attention to the function of the complement system in tumor metastasis in conjunction with the analysis of high-throughput data from the GEO database. Our PCR results revealed that the mRNA levels of C5 were higher in the cell lines and liver metastasis tissues (Figure 4D,4E), and western blot elaborated that C5 was considerably increased in the SW620 and SW480 cell lines compared to the RKO, HT29, and HCT116 cell lines (Figure 4F). Using knockdown technology of C5 in SW480 and SW620 cells, we discovered that C5 could promote the proliferation, migration, and invasion in CRC cell lines (Figures 5,6).
Finally, we collected eight paired LMCRC tissues in our hospital to verify whether the expression of C5 in metastasis tissue was higher than in the primary tumor. The results revealed that C5 was higher in liver metastasis tissue compared with the corresponding CRC (Figure 6). In this study, we employed bioinformatics technology and in vitro experiments to discover a new biomarker for LMCRC, which may provide a new direction for further research in LMCRC. However, there are still issues that need to be resolved. More clinical samples and in vivo experiments are needed to sufficiently validate the results of the current study, and the relevant molecular mechanism needs to be further investigated.
In conclusion, by utilizing three GSE profile datasets and other bioinformatics analyses, our current research found that C5 shows higher expression levels in LMCRC compared with CRC, which may represent a potential clinical target in LMCRC.
Acknowledgments
Funding: This work was funded by the Talent Support Project of Shaanxi Provincial People’s Hospital (No. 2021JY-02), and the National Nature Science Foundation of China (Nos. 82103371, 82102959, 82172739, 81871924, 82072666, 81802893, and 8197282).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-829/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-829/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-829/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Review Board of Zhongshan Hospital of Fudan University (No. B2020-348R) and informed consent was taken from all the patients. Animal experiments were performed under a project license (No. 2021753) granted by the Institutional Animal Care and Use Committee at Fudan University, in compliance with Chinese National Standard (GBT35823-2018) for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lee JK, Merchant SA, Jensen CD, et al. Rising Early-onset Colorectal Cancer Incidence Is Not an Artifact of Increased Screening Colonoscopy Use in a Large, Diverse Healthcare System. Gastroenterology 2022;162:325-327.e3. [Crossref] [PubMed]
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Cloyd JM, Mizuno T, Kawaguchi Y, et al. Comprehensive Complication Index Validates Improved Outcomes Over Time Despite Increased Complexity in 3707 Consecutive Hepatectomies. Ann Surg 2020;271:724-31. [Crossref] [PubMed]
- Kambakamba P, Hoti E, Cremen S, et al. The evolution of surgery for colorectal liver metastases: A persistent challenge to improve survival. Surgery 2021;170:1732-40. [Crossref] [PubMed]
- Rouyer M, François E, Sa Cunha A, et al. Effectiveness of first-line cetuximab in wild-type RAS metastatic colorectal cancer according to tumour BRAF mutation status from the EREBUS cohort. Br J Clin Pharmacol 2021;87:1120-8. [Crossref] [PubMed]
- Dohan A, Gallix B, Guiu B, et al. Early evaluation using a radiomic signature of unresectable hepatic metastases to predict outcome in patients with colorectal cancer treated with FOLFIRI and bevacizumab. Gut 2020;69:531-9. [Crossref] [PubMed]
- Kobayashi S, Takahashi S, Nomura S, et al. BRAF V600E potentially determines "Oncological Resectability" for "Technically Resectable" colorectal liver metastases. Cancer Med 2021;10:6998-7011. [Crossref] [PubMed]
- Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Clin Pract Oncol 2008;5:588-99. [Crossref] [PubMed]
- Wang S, Zhang C, Zhang Z, et al. Transcriptome analysis in primary colorectal cancer tissues from patients with and without liver metastases using next-generation sequencing. Cancer Med 2017;6:1976-87. [Crossref] [PubMed]
- Pan W, Wang K, Li J, et al. Restoring HOXD10 Exhibits Therapeutic Potential for Ameliorating Malignant Progression and 5-Fluorouracil Resistance in Colorectal Cancer. Front Oncol 2021;11:771528. [Crossref] [PubMed]
- Zeng C, Chen Y. HTR1D, TIMP1, SERPINE1, MMP3 and CNR2 affect the survival of patients with colon adenocarcinoma. Oncol Lett 2019;18:2448-54. [Crossref] [PubMed]
- Caramujo-Balseiro S, Faro C, Carvalho L. Metabolic pathways in sporadic colorectal carcinogenesis: A new proposal. Med Hypotheses 2021;148:110512. [Crossref] [PubMed]
- Gu JN, Yao S, Cao YH, et al. Novel parameter based on lipid indicators ratio improves prognostic value of plasma lipid levels in resectable colorectal cancer patients. World J Gastrointest Surg 2021;13:689-701. [Crossref] [PubMed]
- Chen C, Yi W, Zeng ZF, et al. Serum apolipoprotein B to apolipoprotein A-I ratio is an independent predictor of liver metastasis from locally advanced rectal cancer in patients receiving neoadjuvant chemoradiotherapy plus surgery. BMC Cancer 2022;22:7. [Crossref] [PubMed]
- Piao C, Zhang WM, Li TT, et al. Complement 5a stimulates macrophage polarization and contributes to tumor metastases of colon cancer. Exp Cell Res 2018;366:127-38. [Crossref] [PubMed]
- Giannis D, Sideris G, Kakos CD, et al. The role of liver transplantation for colorectal liver metastases: A systematic review and pooled analysis. Transplant Rev (Orlando) 2020;34:100570. [Crossref] [PubMed]
- Stewart CL, Warner S, Ito K, et al. Cytoreduction for colorectal metastases: liver, lung, peritoneum, lymph nodes, bone, brain. When does it palliate, prolong survival, and potentially cure? Curr Probl Surg 2018;55:330-79. [Crossref] [PubMed]
- Wolpin BM, Mayer RJ. Systemic treatment of colorectal cancer. Gastroenterology 2008;134:1296-310. [Crossref] [PubMed]
- Ishaque N, Abba ML, Hauser C, et al. Whole genome sequencing puts forward hypotheses on metastasis evolution and therapy in colorectal cancer. Nat Commun 2018;9:4782. [Crossref] [PubMed]
- Sauter RJ, Sauter M, Reis ES, et al. Functional Relevance of the Anaphylatoxin Receptor C3aR for Platelet Function and Arterial Thrombus Formation Marks an Intersection Point Between Innate Immunity and Thrombosis. Circulation 2018;138:1720-35. [Crossref] [PubMed]
- Ajona D, Ortiz-Espinosa S, Pio R. Complement anaphylatoxins C3a and C5a: Emerging roles in cancer progression and treatment. Semin Cell Dev Biol 2019;85:153-63. [Crossref] [PubMed]
- Afshar-Kharghan V. The role of the complement system in cancer. J Clin Invest 2017;127:780-9. [Crossref] [PubMed]
- Pio R, Ajona D, Ortiz-Espinosa S, et al. Complementing the Cancer-Immunity Cycle. Front Immunol 2019;10:774. [Crossref] [PubMed]
- Akhir FNM, Noor MHM, Leong KWK, et al. An Immunoregulatory Role for Complement Receptors in Murine Models of Breast Cancer. Antibodies (Basel) 2021;10:2. [Crossref] [PubMed]
- Bao D, Zhang C, Li L, et al. Integrative Analysis of Complement System to Prognosis and Immune Infiltrating in Colon Cancer and Gastric Cancer. Front Oncol 2020;10:553297. [Crossref] [PubMed]
- Bondza S, Marosan A, Kara S, et al. Complement-Dependent Activity of CD20-Specific IgG Correlates With Bivalent Antigen Binding and C1q Binding Strength. Front Immunol 2020;11:609941. [Crossref] [PubMed]
- Jackson WD, Gulino A, Fossati-Jimack L, et al. C3 Drives Inflammatory Skin Carcinogenesis Independently of C5. J Invest Dermatol 2021;141:404-414.e6. [Crossref] [PubMed]
- Ding P, Li L, Li L, et al. C5aR1 is a master regulator in Colorectal Tumorigenesis via Immune modulation. Theranostics 2020;10:8619-32. [Crossref] [PubMed]
- Gadwa J, Bickett TE, Darragh LB, et al. Complement C3a and C5a receptor blockade modulates regulatory T cell conversion in head and neck cancer. J Immunother Cancer 2021;9:e002585. [Crossref] [PubMed]
- Daugan MV, Revel M, Russick J, et al. Complement C1s and C4d as Prognostic Biomarkers in Renal Cancer: Emergence of Noncanonical Functions of C1s. Cancer Immunol Res 2021;9:891-908. [Crossref] [PubMed]
- Zheng Z, Li YN, Jia S, et al. Lung mesenchymal stromal cells influenced by Th2 cytokines mobilize neutrophils and facilitate metastasis by producing complement C3. Nat Commun 2021;12:6202. [Crossref] [PubMed]
(English Language Editor: A. Kassem)


