
Identification of characteristic markers correlated with Th2 cell infiltration and metabolism molecular subtype in pancreatic adenocarcinoma
Introduction
Pancreatic adenocarcinoma is one of the deadliest malignancies, with an 8.5% five-year survival rate, making it the third leading cause of cancer-related death (1). Surgical resection is the only curative treatment for pancreatic adenocarcinoma (2). FOLFIRINOX and gemcitabine plus nab-paclitaxel are gradually becoming optional treatment strategies (3), and multiple targeted therapies are being studied. Patients with germline BRCA1 or BRCA2 mutations benefit from poly-ADP ribose polymerase (PARP) inhibitors (4). Clinical trials of CTLA-4 and programmed death ligand-1 (PD-L1) inhibitors are ongoing (5). Chimeric antigen receptor T cells (CAR-T) cell therapy is a potential therapeutic option for pancreatic adenocarcinoma (6). Modulation of novel immune biomarkers may improve the therapeutic outcomes of existing targeted therapies (7).
The T-helper 2 (Th2) cells initiate type 2 immunity and stimulate antibody secretion in response to external stimuli (8). Furthermore, Th2 cell-mediated type 2 immunity influences tumor progression and development (9). The Th2 cells release interleukin (IL)-2 and IL-13 to activate the STAT6 and cMyc pathways and promote pancreatic adenocarcinoma progression (10). This study investigated the application of experimental phenotypes in the identification of novel immune biomarkers.
In recent years, tumor heterogeneity research has served as a bridge between experiments and clinical application. Based on the proteome of hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC), two biomarkers were identified to classify subgroups (11). Triple-negative breast cancer (TNBC) can be classified into four transcriptome-based subtypes with individualized therapeutic strategies (12). Differences in treatment sensitivity have been demonstrated across pancreatic adenocarcinoma subtypes (13). Our research identified the prognostic significance of Th2 cell infiltration, screened three hub genes as characteristic markers correlated with Th2 cell infiltration, and provided a classification for molecular metabolism subtypes in pancreatic adenocarcinoma. We present the following article in accordance with the STREGA reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-333/rc).
Methods
Samples and gene set
We obtained the gene expression matrix and clinical information about of 168 pancreatic adenocarcinoma samples from The Cancer Genome Atlas (TCGA) database. Two samples were deleted because they did not have follow-up data. We also obtained the sample information from the GSE71729 (14), GSE160154 (15), GSE179351 (16), and GSE155698 (17) cohorts in the Gene Expression Ominibus (GEO) database (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) (http://www.ncbi.nlm.nih.gov/geo/).
The landmark cancer genomics program TCGA molecularly characterized over 20,000 primary cancers and matched normal samples spanning 33 cancer types. The working group recruited 186 patients diagnosed with pancreatic adenocarcinoma in 2001–2013, obtained tissue samples with resection or biopsy and collected the follow-up information of 168 patients with the last follow-up time in 2019.
Moffitt et al. (14) analyzed data, including 145 primary and 61 metastatic pancreatic ductal adenocarcinoma (PAAD) samples, 17 cell lines, 46 pancreatic tissue samples, and 88 distant site adjacent normal samples, and collected follow-up information with the last follow-up time in 2015 (GSE71729). Examines expression by paired-end RNA-seq in CD8+ T cells with multiple technical repeats across four biological replicates, with three variables: CAR presence (values: present or absent); antigen exposure (values: control, continuous, other); and time (values: day 0, day 16, day 28) (GSE160154).
In this study, we constructed a metabolism-related gene set (n=4,033) with data downloaded from the Kyoto Encyclopedia of Genes and Genomes (KEGG) (18), Reactome (19), Human genome-scale metabolic models (Human-GEM) (20), and the Braunschweig Enzyme Database (BRENDA) (21). The immune-related gene set (n=2,483) was constructed with data downloaded from the Immport database (https://www.immport.org) and the InnateDB database (https://www.innatedb.ca/). The cellular processes genes set (apoptosis, autophagy, ferroptosis and necroptosis) (n=975) and signal transduction genes set (n=907) were constructed with data from KEGG (18). The gene sets were displayed in tables (website: https://cdn.amegroups.cn/static/public/jgo-22-333-01.zip).
Single sample gene set enrichment analysis (ssGSEA)
The ssGSEA function based on the GSVA package (22) was used to calculate the infiltration scores of 24 types of immune cells (B cells, T cells, T helper cells, Tcm, Tem, Th1 cells, Th2 cells, TFH, Th17 cells, Treg, CD8+ T cells, Tgd, cytotoxic cells, natural killer (NK) cells, NK CD56 dim cells, NK CD56 bright cells, DC, iDC, aDC, pDC , eosinophils, macrophages, mast cells, and neutrophils) for each pancreatic adenocarcinoma sample with the immune cells gene set (website: https://cdn.amegroups.cn/static/public/jgo-22-333-01.zip) (23). We also used ssGSEA to calculate metabolism scores for four types of substances (carbohydrate, lipid, amino acid, and nucleotide) from the metabolism gene sets and to calculate cellular process scores(apoptosis, autophagy, ferroptosis, and necroptosis) from the gene sets (website: https://cdn.amegroups.cn/static/public/jgo-22-333-01.zip). The two gene sets were obtained from KEGG (18).
Weighted gene co-expression network analysis
Weighted gene co-expression network analysis (WGCNA) is a systems biology method used to describe the patterns of the correlation between genes across microarray samples (24). We applied WGCNA to analyze immune, metabolism, cellular processes, and signal transduction gene expression in the TCGA data cohort. Subsequently, gene modules were screened based on the proportion of Th2 cell infiltration. A feature module with the optimal R-squared and P value was selected. Feature genes from the feature module were characterized by module membership (MM) fold value ≥0.8 and gene significance (GS) fold value ≥0.1.
Random survival forest
The R package random survival forest (25) was used to screen hub genes as metabolic markers based on the module feature genes provided by WGCNA. Three hub genes (PLAAT4, TSPOAP1, and MAN2B1) were identified to construct a molecular subtyping expression matrix.
Consensus clustering
Consensus clustering is an unsupervised method to classify samples into subtypes based on feature gene expression (26). The parameters used were: maxK =9, resp=50, pltem =0.8, and pFeature =1. According to the delta area plot, the classification was the most stable at k=2. Moreover, immune cells were classified into two clusters based on infiltration scores.
Drug sensitivity prediction
We selected 13 chemotherapy drugs used to treat pancreatic adenocarcinoma from the Genomics of Drug Sensitivity in Cancer (GDSC; https://www.cancerrxgene.org/) database. The pRRophetic package in R was used to calculate the corresponding IC50 (half maximal inhibitory concentration) (27).
Single nucleotide polymorphism analysis
Single nucleotide polymorphism mutation was analyzed using by the R package maftools (28). The top ten mutation genes were screened, and the mutation type of the ten genes in each pancreatic adenocarcinoma sample was identified. The mutation landscape was displayed as a waterfall plot.
Copy number variation (CNV) analysis
The CNV data was uploaded to GenePattern, and the variation site was estimated with the GISTIC 2.0 module (https://cloud.genepattern.org/). The CNV landscape was obtained from the genome map from the R package maftools (12).
Statistical analysis
The Wilcoxon test or unpaired t-test was used to compare the difference between the two groups. The overall survival (OS) rates were assessed by Kaplan-Meier curves, and differences between survival rates were evaluated with a log-rank test. In the mapping plots, ns shows P>0.05; * shows P≤0.05; ** shows P≤0.01; *** shows P≤0.001; and **** shows P≤0.0001.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Our study is based on open-source data, so there are no ethical issues and other conflicts of interest.
Results
Calculation of immune cell infiltration proportion and identification of prognostic trait immune cells
Firstly, the ssGSEA function from the R package GSVA was used to calculate the immune cell infiltration proportion of pancreatic adenocarcinoma from TCGA-PAAD and GSE71729.Subsequently, ConsensusClusterPlus from the R package was used to classify immune cells into clusters A and B, and the difference in prognosis between immune cells was assessed. The results showed that immune cell infiltration determined the prognosis in pancreatic adenocarcinoma. The interaction network showing infiltration of 24 immune cells is shown in Figure 1A.
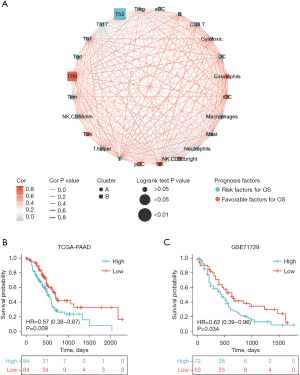
The optimal cut-off value to define high and low Th2 infiltration was determined with the surcut function. Accordingly, pancreatic adenocarcinoma patients were divided into two groups, guaranteeing each group with rational outcome events simultaneously, and the prognosis between the two groups was compared. The group with a low Th2 infiltration proportion displayed a favorable prognosis in TCGA-PAAD and GSE71729 (Figure 1B,1C) (website: https://cdn.amegroups.cn/static/public/jgo-22-333-01.zip).
A previous study had reported that Th2 cells release IL2 and IL13 to activate the STAT6 and cMyc pathways and promote the development of Kras-initiated pancreatic cancer in mice (10). In addition, cellular processes, including apoptosis, ferroptosis, necroptosis (29), and autophagy (30), have been shown to affect the tumor immune microenvironment of pancreatic adenocarcinoma. Multiple signal transduction pathways, including Wnt (31), vascular endothelial growth factor (VEGF) (32), transforming growth factor (TGF)-beta (31), PI3K-Akt (33), nuclear factor (NF)-kappa B (34) and hedgehog (35), have been shown to participate in the growth and metastasis of pancreatic adenocarcinoma and may be associated with immune infiltration. Based on the experimental outcome and prognostic significance, hub genes downstream of Th2 cells were investigated to explore their role in immunity, metabolism, cellular processes and signal transduction. The pancreatic adenocarcinoma subtype classification based on Th2 infiltration was subsequently confirmed.
Construction of a weighted co-expression network and identification of feature hub genes
The 2,483 immune related genes expressed in the matrix in the TCGA-PAAD cohort were searched, and the weighted co-expression network was constructed with the R package WGCNA. Nine gene modules were identified, and the correlation between Th2 infiltration scores and gene modules was calculated. On the bottom right part of each diamond, the color and number represent the P value. The blue module presented the highest correlation coefficient (R=0.53) and the smallest P value (P=2e−13). We used the R package randomForestSRC and randomSurvivalForest to screen the PLAAT4 and TRAF5 genes combined with clinical survival information (Figure 2A). We applied the same procedures to analyze 4,033 metabolism genes, 975 cellular processes genes, and 907 signal transduction genes expression matrices, and compared the variable importance of the screened genes. Three hub genes from the metabolism genes were identified as metabolic markers for subtype classification with Th2 cell infiltration (Figure 2B-2D).
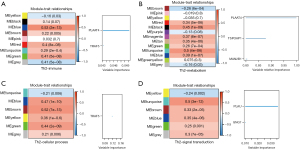
Of the various checkpoint molecules, researchers mainly focused on cytotoxic T lymphocyte associated antigen 4 (CTLA4) and programmed cell death protein 1 (PD1) (36). After blocking CTLA4 and PD1, T cells activated intrinsic metabolism barriers and reduced AKT activation to interfere with glycolysis (37). Furthermore, fatty acid oxidation may be affected by PD1 ligation (38). The three hub genes may also serve as immunotherapy checkpoints. Therefore, we considered exploring pancreatic adenocarcinoma heterogeneity and classifying Th2 infiltration molecular subtypes based on PLAAT4, TSPOAP1, and MAN2B1 gene expression.
Determination and comparative analysis of pancreatic adenocarcinoma metabolism molecular subtype with Th2 cell infiltration based on hub genes
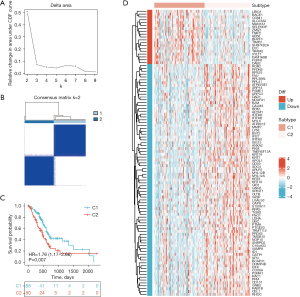
Subsequently, we applied the R package ConsensusClusterPlus with an unsupervised clustering function to analyze the expression matrix of the three hub genes, classifying 168 pancreatic adenocarcinomas into two subtypes (C1 and C2 subtypes; C1 subtype =88 patients, C2 subtype =80 patients) The Delta area plot demonstrated that the classification was reliable and stable when k=2 (Figure 3A,3B). There were significant differences in the prognosis between the C1 and C2 subtypes (Figure 3C). The R package limma was used to screen differentiation expression genes between subtypes, characterized by |logFC|>1 and adj.P value <0.05, and the heat map displayed the expression quantity of the top 100 (the top 20 upregulated and the top 80 downregulated) differentially expressed genes (DEGs) in the subtypes (Figure 3D).
We utilized Wilcoxon test to compare the differences and display the statistical significance in substance metabolism, cell process, and immune cell infiltration between subtypes (Figure 4A). To explore the gene mutation differences between subtypes, the R package maftools was used to analyze the differences of single nucleotide polymorphism (SNP) mutation between subtypes, and the GISTIC_2.0 module on GenePattern to analyze the differences in CNV between subtypes (Figure 4B-4D). Functional enrichment analysis of the DEGs showed that the genes participated in multiple biological processes (Figure 4E,4F).

Previously published research on pancreatic cancer subtyping has emphasized single genetic marker classification, patterns of genomic aberrations, and transcriptome subtypes (13). We observed differences in the prognosis, metabolism of four different substances, cellular processes, immune cell infiltration, SNP, CNV, and DEGs among Th2 metabolic molecular subtypes.
Differentiation comparison of clinical therapy features between the two subtypes
We used the R package pRRophetic to calculate the IC50 levels of six chemotherapy drugs in each pancreatic adenocarcinoma sample and compared the differences between the two subtypes. The IC50 levels of bortezomib, cisplatin, dasatinib, erlotinib, and gefitinib were significantly higher in C1 compared to C2, demonstrating that the C2 subtypes were more sensitive to these chemotherapeutics. In contrast, the IC50 levels of AZD7762 were significantly lower in C1 compared to C2 (Figure 5A-5F). There were evident differences in the Th2 infiltration proportion on day 0 and day 16 in the pancreatic adenocarcinoma samples of patients with CAR-T cell therapy from the GSE160154 cohort (Figure 5G). We used the hub genes expression matrix to classify samples from patients before radiation therapy and before anti-CTLA4 (ipilimumab) and anti-PD1 (nivolumab) antibody treatment from the GSE179351 cohort into subtypes. Subsequently, the Th2 infiltration proportion between subtypes was compared before and after radiotherapy (Figure 5H-5J) (website: https://cdn.amegroups.cn/static/public/jgo-22-333-01.zip).
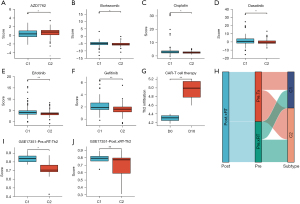
Diagnosis and treatment strategy selection depend on the molecular subtyping classification. Based on Moffitt et al. transcriptomic subtypes, including basal-like and classical subtypes, the basal-like subtype benefits from adjuvant chemotherapy (14). Further, subtype 1 (Hedgehog) and subtype 3 (NOTCH) are targets for immunotherapy, including subtype-specific checkpoint inhibition and myeloid depletion therapy, as described by de Santiago et al. (39). The drug sensitivity to six types of chemotherapeutics between the C1 and C2 subtypes was investigated. Changes in the Th2 infiltration proportion changes from day 0 to day 16 in CAR-T cell therapy before and after radiotherapy were assessed.
Effects of Th2 cell infiltration and metabolism markers in pancreatic adenocarcinoma progression and metastasis
The boxplots displayed the expression of PLAAT4, TSPOAP1, and MAN2B1 between normal pancreatic and pancreatic adenocarcinoma tissues (normal tissues from TCGA and genotype-tissue expression-GTEx, tumor tissues from TCGA) (Figure 6A-6C) (website: https://cdn.amegroups.cn/static/public/jgo-22-333-01.zip). The R package Seurat was applied to analyze the pancreatic adenocarcinoma single-cell RNA sequence data of the GSE155698 cohort. The DimPlot displayed the cell clusters of pancreatic adenocarcinoma (Figure 6D). The FeaturePlot displayed the distribution of the top three upregulated genes and the top three downregulated genes in each pancreatic adenocarcinoma cluster (Figure 6E). There were significant differences in Th2 cell infiltration between normal liver and liver metastatic tissues (Figure 6F-6H) (website: https://cdn.amegroups.cn/static/public/jgo-22-333-01.zip).
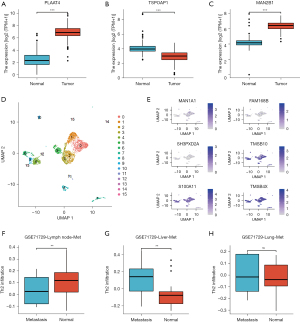
In contrast, in normal and pancreatic adenocarcinoma tissues, we learned that PLAAT4, TSPOAP1, and MAN2B1 expression alongside tumor heterogeneity correlated with tumorigenesis. Various types of immune cell infiltration are involved in cancer metastasis, including macrophages, Treg, and Th17 cells (40). As the above results suggest, Th2 cells may play a role in liver metastasis and the progression of pancreatic adenocarcinoma.
Discussion
A high proportion of Th2 infiltration has been shown to increase the level of IL-4 expressed by the enrichment of basophils in tumor-draining lymph nodes and is related to poor prognosis outcome in pancreatic adenocarcinoma (41). Kinskey et al. discovered that the lymphocyte infiltrate for pancreatic adenocarcinoma had a strong Th2/B cell character and markers of Th2 cells (IL-4 and PTGDR2) were higher expression quantity in pancreatic adenocarcinoma tissue than other cancer tissue (42). Their research provided inspiration for our work, and we speculated a higher Th2 cells infiltration proportion caused high mortality in patients with pancreatic adenocarcinoma. The prognostic value of Th2 cell infiltration was assessed using the immune cell infiltration correlation network. TGF-beta inhibition can block Th2 effector cell activity and is a potential immunotherapeutic approach for other cancers (43). However, research gaps persist about molecular subtyping of pancreatic adenocarcinoma based on Th2 cell infiltration. Our results screened module genes from the immune, metabolism, cellular processes, and signal transduction gene expression matrices and identified optimal hub genes (PLAAT4, TSPOAP1, and MAN2B1) as metabolic markers for subtype classification.
Previous research has typically focused on early single genetic markers to identify genomic aberrations and perform transcriptomic subtyping (44,45). This study classified two subtypes of pancreatic adenocarcinoma with prognostic significance and analyzed the differences between the subtypes (46). Higher levels of fatty acid degradation were observed in the C1 subtype compared to the C2 subtype, which could be the subject of further research. Future studies can also focus on differences in amino acid metabolism, nucleotide metabolism, autophagy, and NK CD56 bright cell infiltration levels. Moreover, we can apply gene detection in clinical practice to identify subtypes based on the mutations.
Only a small minority of pancreatic adenocarcinoma patients benefit from multidisciplinary approaches; the vast majority are insensitive to treatment (3). Researchers can design clinical control trials for subtype heterogeneity treatment based on the differences in subtypes. Treating pancreatic adenocarcinoma with distant metastases is very challenging (47). However, our study results suggest that modulating Th2 cell infiltration could be applied in treating liver metastasis.
We identified the prognostic significance of Th2 cell infiltration in pancreatic adenocarcinoma samples, screened hub genes as characteristic markers to classify subtypes, and compared various aspects between subtypes. A limitation of the study was the lack of clinical practice application results. A pathological immunohistochemical identification system based on hub genes should be established to classify patients with pancreatic adenocarcinoma into subtypes.
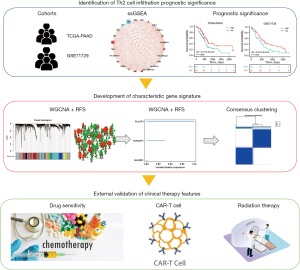
Flow diagram
We drew the flow diagram to display our analysis procedure (Figure 7).
Acknowledgments
Funding: This work was supported by grants from the National Natural Science Foundation of China (Nos. 81871941, 81827807, 81872366). The funding agencies had no role in the study design, data collection and analyses, decision to publish, or preparation of the manuscript.
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-333/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-333/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Our study is based on open-source data, so there are no ethical issues and other conflicts of interest.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin 2022;72:7-33. [Crossref] [PubMed]
- Millikan KW, Deziel DJ, Silverstein JC, et al. Prognostic factors associated with resectable adenocarcinoma of the head of the pancreas. Am Surg 1999;65:618-23; discussion 623-4. [Crossref] [PubMed]
- Mizrahi JD, Surana R, Valle JW, et al. Pancreatic cancer. Lancet 2020;395:2008-20. [Crossref] [PubMed]
- Shroff RT, Hendifar A, McWilliams RR, et al. Rucaparib Monotherapy in Patients With Pancreatic Cancer and a Known Deleterious BRCA Mutation. JCO Precis Oncol 2018; [Crossref] [PubMed]
- Teague A, Lim KH, Wang-Gillam A. Advanced pancreatic adenocarcinoma: a review of current treatment strategies and developing therapies. Ther Adv Med Oncol 2015;7:68-84. [Crossref] [PubMed]
- Stromnes IM, Schmitt TM, Hulbert A, et al. T Cells Engineered against a Native Antigen Can Surmount Immunologic and Physical Barriers to Treat Pancreatic Ductal Adenocarcinoma. Cancer Cell 2015;28:638-52. [Crossref] [PubMed]
- Li TJ, Wang WQ, Yu XJ, et al. Killing the "BAD": Challenges for immunotherapy in pancreatic cancer. Biochim Biophys Acta Rev Cancer 2020;1874:188384. [Crossref] [PubMed]
- Kidd P. Th1/Th2 balance: the hypothesis, its limitations, and implications for health and disease. Altern Med Rev 2003;8:223-46. [PubMed]
- Schreiber S, Hammers CM, Kaasch AJ, et al. Metabolic Interdependency of Th2 Cell-Mediated Type 2 Immunity and the Tumor Microenvironment. Front Immunol 2021;12:632581. [Crossref] [PubMed]
- Dey P, Li J, Zhang J, et al. Oncogenic KRAS-Driven Metabolic Reprogramming in Pancreatic Cancer Cells Utilizes Cytokines from the Tumor Microenvironment. Cancer Discov 2020;10:608-25. [Crossref] [PubMed]
- Gao Q, Zhu H, Dong L, et al. Integrated Proteogenomic Characterization of HBV-Related Hepatocellular Carcinoma. Cell 2019;179:561-577.e22. [Crossref] [PubMed]
- Jiang YZ, Ma D, Suo C, et al. Genomic and Transcriptomic Landscape of Triple-Negative Breast Cancers: Subtypes and Treatment Strategies. Cancer Cell 2019;35:428-440.e5. [Crossref] [PubMed]
- Collisson EA, Bailey P, Chang DK, et al. Molecular subtypes of pancreatic cancer. Nat Rev Gastroenterol Hepatol 2019;16:207-20. [Crossref] [PubMed]
- Moffitt RA, Marayati R, Flate EL, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet 2015;47:1168-78. [Crossref] [PubMed]
- Donahue G. Induction of T cell dysfunction and NK-like T cell differentiation in vitro and in patients after CAR T cell treatment [RNA-seq]; 2020.
- Parikh AR, Szabolcs A, Allen JN, et al. Radiation therapy enhances immunotherapy response in microsatellite stable colorectal and pancreatic adenocarcinoma in a phase II trial. Nat Cancer 2021;2:1124-35. [Crossref] [PubMed]
- Steele NG, Carpenter ES, Kemp SB, et al. Multimodal Mapping of the Tumor and Peripheral Blood Immune Landscape in Human Pancreatic Cancer. Nat Cancer 2020;1:1097-112. [Crossref] [PubMed]
- Kanehisa M, Sato Y, Kawashima M, et al. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 2016;44:D457-62. [Crossref] [PubMed]
- Croft D, O'Kelly G, Wu G, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res 2011;39:D691-7. [Crossref] [PubMed]
- Robinson JL, Kocabaş P, Wang H, et al. An atlas of human metabolism. Sci Signal 2020;13:aaz1482. [Crossref] [PubMed]
- Placzek S, Schomburg I, Chang A, et al. BRENDA in 2017: new perspectives and new tools in BRENDA. Nucleic Acids Res 2017;45:D380-8. [Crossref] [PubMed]
- Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [Crossref] [PubMed]
- Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013;39:782-95. [Crossref] [PubMed]
- Langfelder P, Luo R, Oldham MC, et al. Is my network module preserved and reproducible? PLoS Comput Biol 2011;7:e1001057. [Crossref] [PubMed]
- Strobl C, Boulesteix AL, Kneib T, et al. Conditional variable importance for random forests. BMC Bioinformatics 2008;9:307. [Crossref] [PubMed]
- Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 2010;26:1572-3. [Crossref] [PubMed]
- Geeleher P, Cox N, Huang RS. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One 2014;9:e107468. [Crossref] [PubMed]
- Mayakonda A, Lin DC, Assenov Y, et al. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res 2018;28:1747-56. [Crossref] [PubMed]
- Chen X, Zeh HJ, Kang R, et al. Cell death in pancreatic cancer: from pathogenesis to therapy. Nat Rev Gastroenterol Hepatol 2021;18:804-23. [Crossref] [PubMed]
- Raufi AG, Liguori NR, Carlsen L, et al. Therapeutic Targeting of Autophagy in Pancreatic Ductal Adenocarcinoma. Front Pharmacol 2021;12:751568. [Crossref] [PubMed]
- Geleta B, Tout FS, Lim SC, et al. Targeting Wnt/tenascin C-mediated cross talk between pancreatic cancer cells and stellate cells via activation of the metastasis suppressor NDRG1. J Biol Chem 2022;298:101608. [Crossref] [PubMed]
- Li B, Qin Y, Yu X, et al. Lipid raft involvement in signal transduction in cancer cell survival, cell death and metastasis. Cell Prolif 2022;55:e13167. [Crossref] [PubMed]
- Ouyang L, Liu RD, Lei DQ, et al. MiR-499a-5p promotes 5-FU resistance and the cell proliferation and migration through activating PI3K/Akt signaling by targeting PTEN in pancreatic cancer. Ann Transl Med 2021;9:1798. [Crossref] [PubMed]
- Cykowiak M, Kleszcz R, Kucińska M, et al. Attenuation of Pancreatic Cancer In Vitro and In Vivo via Modulation of Nrf2 and NF-κB Signaling Pathways by Natural Compounds. Cells 2021;10:3556. [Crossref] [PubMed]
- Quatannens D, Verhoeven Y, Van Dam P, et al. Targeting hedgehog signaling in pancreatic ductal adenocarcinoma. Pharmacol Ther 2022;236:108107. [Crossref] [PubMed]
- Parry RV, Chemnitz JM, Frauwirth KA, et al. CTLA-4 and PD-1 receptors inhibit T-cell activation by distinct mechanisms. Mol Cell Biol 2005;25:9543-53. [Crossref] [PubMed]
- Riley JL. PD-1 signaling in primary T cells. Immunol Rev 2009;229:114-25. [Crossref] [PubMed]
- Patsoukis N, Bardhan K, Chatterjee P, et al. PD-1 alters T-cell metabolic reprogramming by inhibiting glycolysis and promoting lipolysis and fatty acid oxidation. Nat Commun 2015;6:6692. [Crossref] [PubMed]
- de Santiago I, Yau C, Heij L, et al. Immunophenotypes of pancreatic ductal adenocarcinoma: Meta-analysis of transcriptional subtypes. Int J Cancer 2019;145:1125-37. [Crossref] [PubMed]
- Kitamura T, Qian BZ, Pollard JW. Immune cell promotion of metastasis. Nat Rev Immunol 2015;15:73-86. [Crossref] [PubMed]
- De Monte L, Wörmann S, Brunetto E, et al. Basophil Recruitment into Tumor-Draining Lymph Nodes Correlates with Th2 Inflammation and Reduced Survival in Pancreatic Cancer Patients. Cancer Res 2016;76:1792-803. [Crossref] [PubMed]
- Kinskey JC, Tu YN, Tong WL, et al. Recovery of Immunoglobulin VJ Recombinations from Pancreatic Cancer Exome Files Strongly Correlates with Reduced Survival. Cancer Microenviron 2018;11:51-9. [Crossref] [PubMed]
- García de Durango CR, Escribese MM, Rosace D. The TGF-β-Th2 axis: A new target for cancer therapy? Allergy 2021;76:3563-5. [Crossref] [PubMed]
- Biankin AV, Kench JG, Colvin EK, et al. Expression of S100A2 calcium-binding protein predicts response to pancreatectomy for pancreatic cancer. Gastroenterology 2009;137:558-68, 568.e1-11.
- Collisson EA, Sadanandam A, Olson P, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med 2011;17:500-3. [Crossref] [PubMed]
- Lee J, Walsh MC, Hoehn KL, et al. Regulator of fatty acid metabolism, acetyl coenzyme a carboxylase 1, controls T cell immunity. J Immunol 2014;192:3190-9. [Crossref] [PubMed]
- Conroy T, Desseigne F, Ychou M, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med 2011;364:1817-25. [Crossref] [PubMed]


