
STING promotes proliferation and induces drug resistance in colorectal cancer by regulating the AMPK-mTOR pathway
Introduction
Colorectal cancer (CRC) is the third most common cancer and the second most common cause of cancer-related deaths (1). According to the latest, most up-to-date GLOBOCAN 2020 estimates, there were 1.9 million new cases of CRC, including anal cancer, and 935,000 CRC-related deaths in 2020 (2). Surgery combined with chemotherapy can substantially ameliorate the prognosis of patients; however, many patients have poor clinical outcomes due to their advanced tumor stage at the time of diagnosis, distant metastasis, or drug resistance. In-depth research on the mechanism of CRC progression will help us to identify novel biomarkers and improve treatments.
Stimulator of interferon genes (STING), which is also, referred to as TMEM173 and STING1, is a key innate immune sensor (3-5). In higher eukaryotes, cyclic guanosine monophosphate-adenosine 5’-monophosphate (AMP) synthase (cGAS)-STING pathway activation has been characterized as an inflammatory mechanism that is induced by cytosolic double-stranded deoxyribonucleic acid (dsDNA) (6). Recent research suggests that STING may serve as an independent prognostic biomarker and potential target for improving anti-cancer immunity in CRC (7). The STING pathway in CRC has not yet been fully elucidated; however, multiple studies suggest that it mediates carcinogenesis (8,9). The rapid proliferation of cancer cells imposes a high energy demand. The AMP-activated protein kinase (AMPK)-mammalian target of the rapamycin (mTOR) pathway plays a vital part in the modification of energy metabolism. The AMPK-mTOR pathway is also related to tumor drug resistance (10,11). STING, which is involved in the regulation of the AMPK-mTOR pathway, has been found in multiple malignant tumors, such as melanoma, gastric cancer, and hepatocellular carcinoma (11-13).
Our previous study demonstrated that fatty acid 2-hydroxylase depletion decreased the chemosensitivity of gastric cancer cells, partially by re-training the AMPK pathway (14). Further, research has shown that Glioma-Associated Oncogene Homolog 1 (GLI1) overexpression, in combination with AKT-mTOR signaling, induces drug resistance in gastric cancer (15). However, further research needs to be conducted to determine whether STING mediates tumor regulation through the AMPK-mTOR pathway and would be a promising therapeutic target in CRC. Our findings may facilitate the assessment of STING as a diagnostic biomarker and characterize the pathway by which STING regulates CRC. We present the following article in accordance with the MDAR reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-957/rc).
Methods
Patient specimens
From 2016 to 2018, 32 pairs of CRC and adjacent tissues were obtained from patients undergoing radical surgery at The First Affiliated Hospital of Soochow University. The tissue specimens were stored in a liquid-nitrogen tank or underwent formalin tissue fixation immediately after resection. The clinicopathological features of the patients were obtained from their electronic medical records. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Biomedical Research Ethics Committee of The First Affiliated Hospital of Soochow University (2021-No:213). We also obtained the written informed consent from the patients and/or their family members. Table 1 sets out the clinicopathological features of the CRC samples.
Table 1
| Clinical parameters | Case no. | STING expression | P value | |
|---|---|---|---|---|
| None or low | High | |||
| Total | 32 | 15 (46.9%) | 17 (53.1%) | |
| Age, years | ||||
| <65 | 14 | 3 (21.4%) | 11 (78.6%) | 0.2656 |
| ≥65 | 18 | 8 (44.4%) | 10 (55.6%) | |
| Gender | ||||
| Male | 19 | 7 (36.8%) | 12 (63.2%) | >0.9999 |
| Female | 13 | 4 (30.8%) | 9 (69.2%) | |
| Tumor size, mm | ||||
| ≤5 | 18 | 7 (38.9%) | 11 (61.1%) | 0.7120 |
| >5 | 14 | 4 (28.6%) | 10 (71.4%) | |
| Tumor location | ||||
| Colon | 12 | 4 (33.3%) | 8 (66.7%) | >0.9999 |
| Rectum | 20 | 7 (35.0%) | 13 (65.0%) | |
| Depth of invasion | ||||
| T1–2 | 4 | 2 (50.0%) | 2 (50.0%) | 0.5932 |
| T3–4 | 28 | 9 (32.1%) | 19 (67.9%) | |
| Lymph node metastasis | ||||
| Yes | 21 | 8 (38.1%) | 13 (61.9%) | 0.0278* |
| No | 11 | 9 (81.8%) | 2 (18.2%) | |
| Degree of differentiation | ||||
| Well | 19 | 7 (36.8%) | 12 (63.2%) | >0.9999 |
| Poor | 13 | 4 (30.8%) | 9 (69.2%) | |
| Metastasis | ||||
| Yes | 8 | 2 (25%) | 6 (75%) | 0.6808 |
| No | 24 | 9 (37.5%) | 15 (62.5%) | |
| Venous or neural invasion | ||||
| Negative | 21 | 9 (42.9%) | 12 (57.1%) | 0.2481 |
| Positive | 11 | 2 (18.2%) | 9 (81.8%) | |
| TNM stage | ||||
| I/II | 21 | 8 (38.1%) | 13 (61.9%) | 0.0278* |
| III/IV | 11 | 9 (81.8%) | 2 (18.2%) | |
*P<0.05. TNM, tumor-node-metastasis.
Cell cultures and transfection
The CRC cell lines were obtained from the Cell Bank of Shanghai (Shanghai, China). After resuscitation, all of the cell lines were sub-cultured for <4 months and cultured in Roswell Park Memorial Institute Medium 1640 or Dulbecco’s Minimal Essential Medium (Thermo Fisher Scientific, Carlsbad, California, USA), which contained 10% fetal bovine serum (FBS, GIBCO) at a temperature of 37 ℃ and a humidity of 5% carbon dioxide (CO2).
For the transfection, PcDNA3.1-Flag-vector and pcDNA3.1-Flag-STING plasmids encoding human wild-type STING were obtained from the Public Protein/Plasmid Library (Nanjing, China). The plasmid sequences were verified via Sanger sequencing. After incubation for 24 hours, the plasmid (50 nm) was transfected into the LoVo cells by Lipofectamine 2000 (Thermo Fisher Scientific) in accordance with the manufacturer's instructions. The subsequent operations were carried out 24–48 hours after the plasmid transfection.
Western blot
Radioimmunoprecipitation assay buffer (Sigma Aldrich) was used to lyse the cells for 30 minutes to extract the total protein (15). Sodium dodecyl-sulfate polyacrylamide gel electrophoresis). was used to isolate the total protein and then transfer the protein to the polyvinylidene fluoride membranes. The bands were blocked with 5% skimmed milk and then incubated with a polyclonal antibody (Cell Signaling Technology) overnight. Next, it was incubated with the secondary antibody, and finally imaged by chemiluminescence. The results were analyzed using ImageJ software (version: 1.4.3, RRID: SCR_003070).
IHC and immunofluorescence staining
The tissues were embedded in paraffin and cut into 5 µm sections. The slices were dewaxed, rehydrated, and blocked with 30% hydrogen peroxide, and then dyed with hematoxylin and rinsed with tap water. Next, 0.5% hydrochloric acid ethanol solution was added into the slices. The samples were soaked for several seconds and then rinsed with tap water. Next, the samples were stained with eosin solution. The samples were dehydrated with ethanol and xylene. The tissues were sealed with neutral resin. After blocking with goat serum, the sections were incubated with antibodies (Cell Signaling Technology, USA; 1:200 dilution), and then rinsed and incubated with secondary antibodies for 30 minutes. And the results are calculated by the color intensity and the number of positive cells (14). The proportion fraction of positive cells (14). The proportion fractions of the positive cells in the immunohistochemical sections were as follows: 0: <5%; 1: 5–25%; 2: 25–50%; 3: 50–75%; and 4: >75%. multiplied by dye intensity fraction; The degree of staining was divided into negative (0–1), weak positive (2–3), positive (4–7), and strong positive (8–12). In this study, immune response scores of 0–4 and 5–12 were considered negative and positive, respectively.
Bioinformatics analysis
GSE100179 data set is based on the GPL17586 platform (HTA-2_0) Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version]. The GSE100179 data set comprised 20 pairs of non-cancerous and CRC tissues. The gene expression data of the 40 samples were used for the in-depth analysis. The Cancer Genome Atlas (TCGA) of UALCAN (http://ualcan.path.uab.edu/) was also used for the in-depth analysis. The correlations between STING messenger ribonucleic acid (RNA) expression in colon adenocarcinoma (COAD) and individual tumor stage and lymph node metastasis were analyzed. The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) tools were used for the gene annotation. In this study, we used Metascape to analyze the genes that interact with STING. We also used the STRING online database (https://string-db.org/) as a search tool to identify the interacting genes. TCGA Pan-Cancer Atlas data set was used to explore the co-expression of "STING" and "mTOR." The competing endogenous RNA (ceRNA), the TCGA, pancancer Atlas dataset was choosed. CeRNA network comprised coding RNA and non-coding RNA. First, we used TargetScan and ENCORI (The Encyclopedia of RNA Interactomes, http://starbase.sysu.edu.cn/index.php) to predict all the STING miRNAs (microRNAs). Second, we searched for the miRNAs in COAD on ENCORI. Third, we used LncBase v.2 to predict the associated long non-coding RNAs (Precision-Recall (PR) score >0.8). Finally, a ceRNA network was constructed using Cytoscape software. TIMER (https://cistrome.shinyapps.io/timer/) is a website mainly related to immune penetration abundance, which is calculated by various immune deconvolution methods. We retrieved the “STING; COAD” from the in “Gene module” and performed an immune infiltration analysis. We also analyzed the expression data of STING in many tumors. All the gene expression data were obtained from UCSC Xena. R-package “ggpubr” was used to generate all the charts
6-carboxy-FAM-labeled siRNA uptake
The cells were cultured in 96-well plates for 24 hours. Transfection reagent (RNAiMAX/liposome™ 2000) was added to the medium with 5, 10, or 20 nm of phosphoramidate (FAM)-labeled negative control small-interfering RNA (FAM siRNA). Next, use 100 µL Opti-Minimal Essential Medium (MEM) (1x), glutamine and 5% FBS replace the medium, and then add Lipofectamine RNAiMAX reagent to the dish. Reagents and different concentrations of FAM siRNA were diluted in serum-free Opti-MEM at a ratio of 1:1. When the cells were cultured for 5 hours and 24 hours incubation, the cells were washed with phosphate buffer solution once respectively. We used Hoechst 33342 (Cell Signaling Technology) fluorescent dye to co culture with cells for 15 minutes to make the nucleus fluorescent. ImageJ software was used to analyze the image by background subtraction.
STING silencing
The synthetic siRNA targeting human STING (specific target sequence: 5'-GCAUCAAGGAUCGGGUUU-3') comes from IBSBIO. An interfering siRNA (5'-UUCUCCGAACGUGUCACGUTT-3') was used as a negative control. Before the experiment, the cells were inoculated in 6-well dishes with (2.5–3)×105 cellsper well and cultured in their respective medium for 24 hours. The CRC cells were transfected using Lipofectamine rnaimax (Thermo Fisher Scientific) reagent as described previously (14).
Cell viability
The Cell Counting Kit-8 (CCK-8; Donjindo, Japan) was used to measure cell viability. The cells were cultured in 96 well dishes. After 24 hours of incubation, the supernatant from each well was removed. CCK-8 solution and cell culture medium was added in accordance with the manufacturer’s instructions. Finally, the absorbance was detected for 5 consecutive days using a multifunctional microplate reader. Each operation was carried out in triplicate.
Transwell invasion and migration assays
Transwell plates (Corning Incorporated, USA) were used for the subsequent experiments. The matrix gel used for the invasion test was slowly thawed on ice at 4 ℃. The chamber insert used for the invasion analysis was coated with a dilute matrix gel and dried at 37 ℃. The cells (1×105) were placed in the upper chamber of each incubator, and suspended in serum-free medium. 20% FBS intact medium was added to the inferior chamber to induce the cells to move downward. After 24 hours of incubation, the cells were fixed and stained. Finally, the transitional cells were quantified using an inverted microscope (Nikon).
Glucose-uptake assays
After the siRNA transfections, the cells were treated with 0.1 mM of 2-NBDG (2-(N-(7-Nitrobenz-2-oxa-1, 3-diazol-4-yl) Amino)-2-Deoxyglucose) (Invitrogen) in a culture medium. The plates were incubated at 37 ℃ with 5% CO2 for a set period as described above. The unified acquisition settings on the fluorescence microscope (Leica) were used to obtain images. The average fluorescence intensity was analyzed by image J.
Statistical analysis
The data are presented as the mean ± standard deviation. GraphPad Prism 8.0 (GraphPad Software, Inc.) was used for the statistical analysis. The χ2 or Fisher’s exact test was used to evaluate the relationships between the categorical clinicopathological variables and the STING expression levels. A 2-tailed Student’s t-test was used to compare the experimental groups. A 2-way analysis of variance and Sidak’s post-hoc test were used to compare the groups. A value P<0.05 indicated a statistically significant difference.
Results
In CRC, STING expression is upregulated and correlated with advanced tumor stage and a poor prognosis
The analysis of the COAD Gene Expression Omnibus (GEO) data set (GSE100179) and TCGA data set revealed that the expression level of STING was significantly increased in the CRC tissues (Figure 1A,1B).The UALCAN results showed that the messenger RNA (mRNA) of STING was particularly high in patients with a high histological grade (poor differentiation) and a high clinical stage (Figure 1C,1D). The immunohistochemistry (IHC) staining results revealed that STING was expressed in 17 of the 32 tumor samples and 4 of the 32 paired adjacent tissues (Figure 1E-1G). Furthermore, the expression level of STING was related to whether the patients had lymph node metastasis. The STING expression of the CRC patients with lymph node metastasis was significantly increased (Figure 1H, P<0.01). Both the Western blot and IHC staining results demonstrated that STING was more highly expressed in the CRC tissues than the matched surrounding normal tissues (Figure 1I,1J).
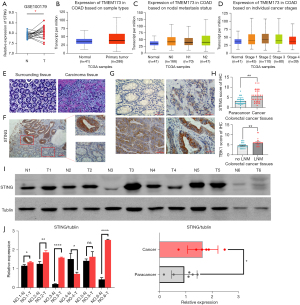
The expression level of STING was related to the clinicopathological indexes of patients with CRC
The STING expression levels were classified according to the IHC staining scores and related to the clinicopathological indicators. STING expression was associated with lymphatic metastasis (P<0.05) and tumor, node, metastasis (TNM) stage (P<0.05). However, no significant associations were found among the following parameters: age, sex, tumor diameter, depth of tumor invasion, and tumor differentiation (Table 1). The P values of each group were >0.05, and while there was no statistical difference between the groups, the number of samples and the effect of the cut-off value for STING expression may have limited the statistical power of parameters: age, sex, tumor diameter, depth of tumor invasion, and tumor differentiation.
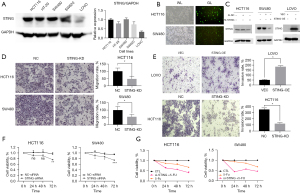
Effects of targeting STING expression on the proliferation, migration, invasion, and drug sensitivity of the CRC cell lines
Western blotting was used to detect STING protein expression in all 5 CRC cell lines. The expression levels were elevated in all the CRC cell lines except LoVo. The expression levels of the HCT116 and SW480 lines were similar (Figure 2A). Transfecting the HCT116 and SW480 cells with STING-siRNA (Figure 2B) significantly reduced STING expression (Figure 2C). The number of migrating and invading cells in the STING-knockdown (STING-KD) group was significantly lower than that in the control group (Figure 2D,2E, P<0.05). Further, different expression levels of STING affected the migration and invasion of CRC cells. The CCK-8 results confirmed that the cell proliferation of the STING-KD group was significantly more decreased at 72 hours compared to that of the untreated group and the negative control group (NC-siRNA) (Figure 2F, P<0.05). These results suggest that elevated STING expression may promote CRC cell proliferation. Further, the CCK-8 assays also demonstrated that the CRC cells in the STING-siRNA group were highly sensitive to 72 hours of 5-FU exposure (Figure 2G, P<0.05).
The effects of STING on CRC cells are mediated by the AMPK-mTOR pathway, and STING regulates glucose uptake in CRC cells
We conducted a number of bioinformatics analyses, including co-expression, GO, KEGG and protein-protein interaction (PPI) analyses, to examine the potential functions of STING in CRC. First, using the STRING database, a PPI network comprising 21 proteins was established (Figure 3A). Next, a GO/KEGG enrichment analysis of the 21 genes was conducted, and the results showed that STING was involved in a series of biological processes in CRC (Figure 3B,3C). The cBioPortal outcomes indicated that STING is related to the mTOR in CRC (Figure 3D). Combined with the above results, we hypothesized that STING promotes the progress of CRC through the mTOR pathway. Transfecting the HCT116 and SW480 cells with STING-siRNA inhibited STING expression (Figure 3E). Additionally, the expression levels of AMPK, p-AMPK, mTOR, and p-mTOR were detected (Figure 3F). Western blot results showed that STING knockdown significantly inhibited AMPK-mTOR signaling, which provided further evidence of the regulatory effects of STING. Thus, our results showed that STING promotes the occurrence and development of CRC cells by inhibiting AMPK and activating the mTOR-related pathway.
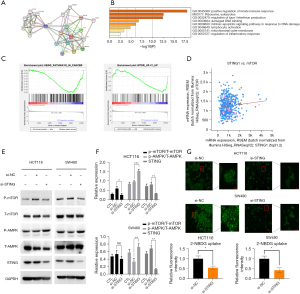
Our experimental results provided preliminary evidence that STING regulates the migration, proliferation, invasiveness, and drug sensitivity of CRC cells by mediating the AMPK-mTOR pathway, which in turn regulates a series of important functions, such as intracellular energy metabolism and biosynthesis. AMPK and mTOR are intracellular energy receptors that balance the production, consumption, and synthesis of intracellular energy sources through negative feedback regulation to maintain homeostasis. Additionally, mTOR mediates some downstream signaling pathways and is involved in the metabolism of sugars, lipids, amino acids, and other substances. We examined whether the increased glucose uptake observed in the tumor cells was regulated by STING signaling, and found that the glucose uptake of the STING knockdown group was significantly lower than that of the control group (Figure 3G). This suggests that STING is not only involved in regulating AMPK-mTOR pathway activity, but also regulates glucose uptake, which might further mediate CRC cell proliferation and metastasis.
hsa-miR-193b-3p may be a key miRNA in the ceRNA network
Based on the prediction results of TargetScan and ENCORI, we identified 20 miRNAs (number of repetitions ≥7). We then used lncbase v.2 (PR score >0.8) to predict the 105 lncRNAs related to these miRNAs. Next, we constructed a STING ceRNA network using Cytoscape (Figure 4A,4B). A search of the 20 miRNAs on ENCORI showed that in terms of the differential expression and the survival rate, only has-mir-193b-3p had obvious clinical significance (Figure 4C,4D).
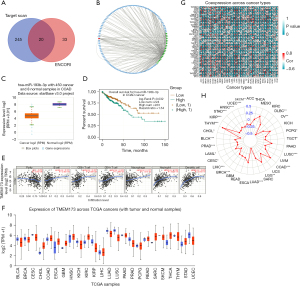
STING affects the infiltration of immune cells in COAD and is a key factor in many tumors
The TIMER results confirmed that in COAD, STING was related to B cells, cluster of differentiation (CD)4+ T cells, CD8+ T cells, macrophages, neutrophils and, mDC cells in the infiltrating tumor tissues (Figure 4E). Thus, our findings that STING affects the progression of CRC by regulating immune cell infiltration represents a promising research direction. Further, our Pan-Cancer analysis showed that STING was potentially a common oncogene in many tumors (Figure 4F-4H).
Discussion
The tumorigenesis of gastrointestinal malignancies is closely related to non-specific inflammation resulting from the aberrant activation of the innate immune system (16). STING dysfunction may cause CRC and increase the susceptibility of melanoma cells to oncolytic viruses (17,18). STING dysfunction may cause CRC and increase the susceptibility of melanoma cells to oncolytic viruses (17,19), thus, STING may serve as an adjunctive therapeutic target for CRC. Further, human papillomavirus E7 and adenovirus E1A oncoproteins bind to STING and inhibit its function (20), which suggests that STING, to some extent, plays a protective role against infections caused by these carcinogenic viruses.
STING expression is reduced in gastric cancer, and decreased levels of STING are associated with a poor prognosis (21). Our TCGA database analysis confirmed that the expression of STING is elevated in CRC patients. In an earlier study, STING served as a biomarker for overall survival after adjusting for tumor stage and intratumoral CD8+ T-cell infiltration (7). Further, STING expression was shown to be upregulated in a consensus molecular subgroup-1 of CRC patients (6,22). Thus, we hypothesized that elevated STING expression in CRC tissues indicates a poor prognosis. To test our hypothesis, we evaluated STING expression levels in multiple patient-derived CRC tissue specimens and corresponding adjacent normal tissues, and found that STING expression was elevated in the CRC tissues. We also examined the correlation between STING expression levels and clinical indicators, and found that high STING expression levels were related to advanced TNM stages of CRC.
Previous studies have shown that STING-mediated immune pathways are involved in the tumorigenesis of many malignancies. Abnormal STING function significantly affects cancer cell proliferation, metastasis, and anti-tumor immunity (18). Tumor drug sensitivity is enhanced by the recruitment and infiltration of immune effector cells stimulated by STING-mediated interferon (IFN) production triggered by increased cytoplasmic dsDNA induced by adjuvant chemotherapy (18,23-26). However, further research needs to be conducted on the potential roles of STING-related pathways as therapeutic targets that inhibit tumorigenesis, enhance anti-tumor immunity, and reduce drug resistance in CRC (27,28). And the sensing of cytoplasmic micronucleus (the product of faulty segregation of damaged chromosomes during mitosis) is considered to be the key event linking genotoxic stress with aging phenotype (including SASP) by cGAS and STING. Interferon-β (IFN β) is produced in cells with DNA damage, CGAS-STING signal has been proved to be effective in IRF3 driven IFN β induction in some but not all conditions promoting cellular senescence (29).
In the present study, we selected HCT116 and SW480 CRC cell lines with high STING expression levels and then downregulated STING expression by siRNA interference. We then used the constructed cell model to perform relevant cell function experiments. First, we found that CRC cell proliferation was significantly decreased after STING knockdown, which supported our hypothesis that elevated STING expression promotes CRC cell proliferation. We also confirmed these results by conducting clonogenic assays. In addition, our transwell assays indicated that STING knockdown reduced CRC cellular migration and invasiveness. Taken together, our results suggest that STING signaling significantly promotes CRC development.
STING signaling may also affect CRC drug resistance (22,30). Our study suggests that STING knockdown improves the chemosensitivity of CRC cells to 5-FU. Notably, 5-FU activates the cGAS-STING pathway that produces the type I IFN in CRC cells. Conversely, Tian et al. reported that STING did not significantly change the lethal effect of 5-FU on colon cancer cells in vitro. Tian et al. speculated that endogenous IFN expression by cancer cells was insufficient to achieve enhanced cytotoxicity in vitro (30).
STING-mediated signaling also inhibits tumor growth by activating the innate immune system, thereby upregulating the downstream secretion of immune regulatory factors, such as IFN, and the consequent recruitment and infiltration of immune effector cells, including macrophages, dendritic cells, and CD8+ lymphocytes (18,19,22). However, functional defects in STING-related pathways may inhibit IFN secretion in CRC cell lines (31-34). A recent study confirmed that CGAs prevents colon cancer (35). Recently, many studies have shown that activating STING and stimulating the production of type I interferon are very important for anti-cancer immune response (36,37). In addition, new research also shows that STING can also regulate anti-cancer immunity in a way independent of type I interferon. For example, STING activation has been proven to enhance the presentation of cancer antigens, help start and activate T cells, promote T cell trafficking and infiltration into tumors, and promote T cells to recognize and kill cancer cells (28).
Our study showed that STING overexpression was associated with markers of advanced CRC. We knocked down STING in the CRC cell line and found that reduced STING expression was accompanied by decreased proliferation and migration and enhanced 5-FU sensitivity. However, STING-related signaling is often impaired in CRC. Other studies have shown that STING-mediated downstream antitumor immune responses activated by DNA damage enhance radiation- and chemotherapy-induced cytotoxicity (7,26,38). In view of the above results, we hypothesized that STING regulates the function of CRC cells through an IFN-independent pathway.
In this study, we used GEO microarray data for the gene set enrichment analysis (GSEA) to identify the pathways that were significantly enriched after STING activation. We then focused on changes in mTOR-related pathway function. Combined with our previous findings (14,15,39), our present findings demonstrate that STING activation upregulated the mTOR-related pathway. Research has shown that mTOR activation promotes intracellular metabolism, enhances protein synthesis, inhibits autophagy, and forms a negative feedback loop with AMPK (9). Thus, it appears that STING regulates CRC cells through the AMPK-mTOR pathway. We found that AMPK function increased after STING downregulation, while mTOR function decreased. This suggests that a series of high energy-consuming processes are inhibited in CRC cells (40). Additionally, the effect of STING signaling on glucose uptake in the CRC cells was explored. The glucose uptake rate decreased with the downregulation of STING, which provides further evidence that STING regulates energy metabolism. Thus, the above results show that the regulation of STING is mediated by the AMPK-mTOR pathway, which may alter energy metabolism and affect proliferation, migration, and drug sensitivity.
The present study had some limitations. First, we only briefly described the effect of the mTOR pathway on cell energy homeostasis. Second, while glucose entry into cells is mediated by membrane transporters, the correlation between mTOR pathway and glucose uptake appears to be very small. However, it should be noted that the mTOR pathway is closely related to autophagy. A variety of organelle membranes are involved in autophagy, and the renewal of various organelle membranes is inseparable from cell membranes. The renewal of cell membranes must involve dynamic changes in cell membrane proteins. More importantly, there is also an important link between autophagy and energy metabolism. There is no doubt that the most important mechanism of cell metabolism is glucose metabolism. In the future, we intend to explore this mechanism further. Notably, in vivo experiments and experiments that monitor important CRC-related indicators need to be conducted. And, we boldly put forward the following ideas on how to further treat CRC through STING. STING inhibitors can cooperate with traditional chemotherapy drugs to trigger anti-tumor response; STING inhibitor combined with tumor vaccine may be applied to human tumor therapy.
Conclusions
In this study, we identified a novel mechanism that links STING to the AMPK-mTOR pathway. This mechanism can be used to monitor the progression of CRC, identify therapeutic targets, and thus improve clinical outcomes. We also reverse constructed a STING ceRNA network in COAD and identified a key miRNA that can be used in further in-depth research.
Acknowledgments
Funding: This work was supported by the Key Laboratory Project of the Clinical Pharmacy of Jiangsu Province of China (No. XZSYSKF2020027 to Guoqiang Zhou), the Gusu Medical Key Talent Project of Suzhou City of China (No. GSWS2020005 to Songbing He), the New Pharmaceutics and Medical Apparatuses Project of Suzhou City of China (No. SLJ2021007 to Songbing He), the Science and Technology Development Plan Project of Suzhou City of China (No. SYS2019007 to Guoqiang Zhou), and the Clinical Medical Expert Team Project of Suzhou City of China (No. CSYJTD202101 to Jinbing Sun).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-957/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-957/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-957/coif). The authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zhou E, Rifkin S. Colorectal Cancer and Diet: Risk Versus Prevention, Is Diet an Intervention? Gastroenterol Clin North Am 2021;50:101-11. [Crossref] [PubMed]
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 2009;461:788-92. [Crossref] [PubMed]
- Rivera Vargas T, Benoit-Lizon I, Apetoh L. Rationale for stimulator of interferon genes-targeted cancer immunotherapy. Eur J Cancer 2017;75:86-97. [Crossref] [PubMed]
- Corrales L, Gajewski TF. Molecular Pathways: Targeting the Stimulator of Interferon Genes (STING) in the Immunotherapy of Cancer. Clin Cancer Res 2015;21:4774-9. [Crossref] [PubMed]
- Vashi N, Bakhoum SF. The Evolution of STING Signaling and Its Involvement in Cancer. Trends Biochem Sci 2021;46:446-60. [Crossref] [PubMed]
- Chon HJ, Kim H, Noh JH, et al. STING signaling is a potential immunotherapeutic target in colorectal cancer. J Cancer 2019;10:4932-8. [Crossref] [PubMed]
- Marill J, Mohamed Anesary N, Paris S. DNA damage enhancement by radiotherapy-activated hafnium oxide nanoparticles improves cGAS-STING pathway activation in human colorectal cancer cells. Radiother Oncol 2019;141:262-6. [Crossref] [PubMed]
- Jiang X, Liu G, Hu Z, et al. cGAMP inhibits tumor growth in colorectal cancer metastasis through the STING/STAT3 axis in a zebrafish xenograft model. Fish Shellfish Immunol 2019;95:220-6. [Crossref] [PubMed]
- Inoki K, Kim J, Guan KL. AMPK and mTOR in cellular energy homeostasis and drug targets. Annu Rev Pharmacol Toxicol 2012;52:381-400. [Crossref] [PubMed]
- Wang F, Alain T, Szretter KJ, et al. S6K-STING interaction regulates cytosolic DNA-mediated activation of the transcription factor IRF3. Nat Immunol 2016;17:514-22. [Crossref] [PubMed]
- Prantner D, Perkins DJ, Vogel SN. AMP-activated Kinase (AMPK) Promotes Innate Immunity and Antiviral Defense through Modulation of Stimulator of Interferon Genes (STING) Signaling. J Biol Chem 2017;292:292-304. [Crossref] [PubMed]
- Moretti J, Blander JM. Detection of a vita-PAMP STINGs cells into reticulophagy. Autophagy 2018;14:1102-4. [Crossref] [PubMed]
- Yao Y, Yang X, Sun L, et al. Fatty acid 2-hydroxylation inhibits tumor growth and increases sensitivity to cisplatin in gastric cancer. EBioMedicine 2019;41:256-67. [Crossref] [PubMed]
- Yao Y, Zhou D, Shi D, et al. GLI1 overexpression promotes gastric cancer cell proliferation and migration and induces drug resistance by combining with the AKT-mTOR pathway. Biomed Pharmacother 2019;111:993-1004. [Crossref] [PubMed]
- Terzić J, Grivennikov S, Karin E, et al. Inflammation and colon cancer. Gastroenterology 2010;138:2101-2114.e5. [Crossref] [PubMed]
- Xia T, Konno H, Barber GN. Recurrent Loss of STING Signaling in Melanoma Correlates with Susceptibility to Viral Oncolysis. Cancer Res 2016;76:6747-59. [Crossref] [PubMed]
- Corrales L, Glickman LH, McWhirter SM, et al. Direct Activation of STING in the Tumor Microenvironment Leads to Potent and Systemic Tumor Regression and Immunity. Cell Rep 2015;11:1018-30. [Crossref] [PubMed]
- Xia T, Konno H, Ahn J, et al. Deregulation of STING Signaling in Colorectal Carcinoma Constrains DNA Damage Responses and Correlates With Tumorigenesis. Cell Rep 2016;14:282-97. [Crossref] [PubMed]
- Lau L, Gray EE, Brunette RL, et al. DNA tumor virus oncogenes antagonize the cGAS-STING DNA-sensing pathway. Science 2015;350:568-71. [Crossref] [PubMed]
- Song S, Peng P, Tang Z, et al. Decreased expression of STING predicts poor prognosis in patients with gastric cancer. Sci Rep 2017;7:39858. [Crossref] [PubMed]
- Chen SY, Chen S, Feng W, et al. A STING-related prognostic score predicts high risk patients of colorectal cancer and provides insights into immunotherapy. Ann Transl Med 2021;9:14. [Crossref] [PubMed]
- Woo SR, Fuertes MB, Corrales L, et al. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity 2014;41:830-42. [Crossref] [PubMed]
- Deng L, Liang H, Xu M, et al. STING-Dependent Cytosolic DNA Sensing Promotes Radiation-Induced Type I Interferon-Dependent Antitumor Immunity in Immunogenic Tumors. Immunity 2014;41:843-52. [Crossref] [PubMed]
- Demaria O, De Gassart A, Coso S, et al. STING activation of tumor endothelial cells initiates spontaneous and therapeutic antitumor immunity. Proc Natl Acad Sci U S A 2015;112:15408-13. [Crossref] [PubMed]
- Ahn J, Xia T, Konno H, et al. Inflammation-driven carcinogenesis is mediated through STING. Nat Commun 2014;5:5166. [Crossref] [PubMed]
- Yi G, Brendel VP, Shu C, et al. Single nucleotide polymorphisms of human STING can affect innate immune response to cyclic dinucleotides. PLoS One 2013;8:e77846. [Crossref] [PubMed]
- Cerboni S, Jeremiah N, Gentili M, et al. Intrinsic antiproliferative activity of the innate sensor STING in T lymphocytes. J Exp Med 2017;214:1769-85. [Crossref] [PubMed]
- Chavanet A, Hill KR, Jiménez-Andrade Y, et al. Intracellular signaling modules linking DNA damage to secretome changes in senescent melanoma cells. Melanoma Res 2020;30:336-47. [Crossref] [PubMed]
- Tian J, Zhang D, Kurbatov V, et al. 5-Fluorouracil efficacy requires anti-tumor immunity triggered by cancer-cell-intrinsic STING. EMBO J 2021;40:e106065. [Crossref] [PubMed]
- Pépin G, Gantier MP. Assessing the cGAS-cGAMP-STING Activity of Cancer Cells. Methods Mol Biol 2018;1725:257-66. [Crossref] [PubMed]
- Vander Heiden MG, DeBerardinis RJ. Understanding the Intersections between Metabolism and Cancer Biology. Cell 2017;168:657-69. [Crossref] [PubMed]
- Stine ZE, Walton ZE, Altman BJ, et al. MYC, Metabolism, and Cancer. Cancer Discov 2015;5:1024-39. [Crossref] [PubMed]
- Li Z, Zhang H. Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cell Mol Life Sci 2016;73:377-92. [Crossref] [PubMed]
- Hu S, Fang Y, Chen X, et al. cGAS restricts colon cancer development by protecting intestinal barrier integrity. Proc Natl Acad Sci U S A 2021;118:e2105747118. [Crossref] [PubMed]
- Galluzzi L, Vanpouille-Box C, Bakhoum SF, et al. SnapShot: CGAS-STING Signaling. Cell 2018;173:276.e1. [Crossref] [PubMed]
- Pantelidou C, Sonzogni O, De Oliveria TM, et al. PARP inhibitor efficacy depends on CD8(+) T-cell recruitment via Intratumoral STING pathway activation in BRCA-deficient models of triplenegative breast Cancer. Cancer Discov. 2019;9:722-37. [Crossref] [PubMed]
- Sprooten J, Agostinis P, Garg AD. Type I interferons and dendritic cells in cancer immunotherapy. Int Rev Cell Mol Biol 2019;348:217-62. [Crossref] [PubMed]
- Lu T, Sun L, Zhu X. Yes-associated protein enhances proliferation and attenuates sensitivity to cisplatin in human gastric cancer cells. Biomed Pharmacother 2018;105:1269-75. [Crossref] [PubMed]
- Kim LC, Cook RS, Chen J. mTORC1 and mTORC2 in cancer and the tumor microenvironment. Oncogene 2017;36:2191-201. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


