
Interfering HMGB3 release from cancer-associated fibroblasts by miR-200b represses chemoresistance and epithelial-mesenchymal transition of gastric cancer cells
Introduction
Gastric cancer (GC) is the most common tumor of the digestive system. Although the incidence rate of GC has decreased in recent years, the overall mortality rate remains the third highest in the world. The incidence of GC is mainly concentrated in the Western Pacific region or developing countries, and especially in Japan and China (1,2). At present, partial or total gastrectomy is the preferred treatment for GC, followed by chemotherapy. However, the five-year survival rate of GC patients is still only 4% (3). Meanwhile, post-treatment complications such as dumping syndrome, anastomotic fistula, infection, chemoresistance, and hair loss seriously affect the patient quality of life. Molecule-targeted therapy has been shown to enhance the therapeutic effect of malignant tumors.
Fibroblasts contribute to maintaining skin integrity, but normal fibroblasts can be activated into cancer-associated fibroblasts (CAFs) in the tumor microenvironment (TEM) (4). A study has shown that CAFs directly interact with cancer cells, change the microenvironment, control the immune reaction, deposit various extracellular matrix (ECM) components, and stimulate angiogenesis by regulating paracrine signaling through inflammatory factors, thus promoting tumor metastasis and invasion (5). Zhang et al. found that ovarian cancer cells and TEM activate CAFs, and that gold nanoparticles exert tumor-suppressive effects by inactivating CAFs (6). Other researchers have found that CAFs activate the signal transduction of JAK2/STAT3 by secreting interleukin (IL)-17a, thereby enhancing GC cell migration and invasion (7). These findings indicate that CAFs have great potential in cancer treatment.
MicroRNAs (miRNAs) are non-coding small RNA with a length of 21 to 23 nucleotides, which regulate the target messenger RNAs (mRNAs) after transcription through translation inhibition and degradation promotion (8). A recent study has shown that miRNAs are crucial for the diagnosis, treatment, and prognosis of cancer. According to Tsai et al., miR-122, miR-195-5p, miR-203, miR-218, and miR-375 are down-regulated in GC and are diagnostic biomarkers. Overexpressed miR-10b, miR-21, and miR-212 can be used as clinical indicators of high-risk metastasis and poor prognosis. Overexpression of miR-1207-5p and miR-1266 restrains GC growth, which can be used to develop new treatment strategies (9). A study has shown that the miR-200 family can not only restrain tumor epithelial-mesenchymal transition (EMT) and metastasis, inhibit the self-renewal and differentiation of cancer stem cells, but also reverse chemoresistance (10). MiR-200b is an important member of the miR-200 family. One study has revealed that miR-200b abates the growth and chemoresistance of lung cancer by targeting p70S6K1 to interrupt the AKT and ERK1/2 signal transduction (11). However, the influence of miR-200b on the microenvironment of CAF-GC cells remains to be further studied.
High Mobility Group Box (HMGB) family have 3 vital members, including HMGB1 (12), HMGB2 (13), HMGB3 (14), are all involved in cancer development and progression, and have prognostic and immunological values (15). Interestingly, HMGB1, HMGB2, and HMGB3 expression was higher in tumor tissues than in normal tissues, especially in GC. High HMGB1, HMGB2, and HMGB3 expression may predict a poor prognosis among patients with GC (16). HMGB3 (HMGB2a) is featured by two box domains (boxes A and B) and acidic C-terminal tails. HMGB3 contributes to DNA repair, recombination, transcription, and replication (17). Nemeth et al. found that mice lacking HMGB3 have more stable self-renewal and differentiation of hematopoietic stem cells, which is closely related to the activation of the Wnt signaling pathway (18). However, some researchers have argued that inhibition of HMGB3 reduces the proliferation and migration of trophoblast cells and promotes apoptosis, contributing to the amelioration of fetal growth restriction (19). In addition, HMGB3 exerts a vital role in tumors. It has been demonstrated that HMGB3 activates WNT/β-catenin, thereby promoting colorectal cancer (CRC) cell proliferation and migration (20). Mukherjee et al. have shown that inhibiting HMGB3 enhances the sensitivity of ovarian cancer cells to cisplatin (CDDP) (21). Another study has manifested that HMGB3 is highly expressed in gastric adenocarcinoma and promotes cell proliferation, which is closely related to poor prognosis (22).
In the present study, the primary objective was to evaluate the influence of HMGB on CAF-related GC (GC-CAFs), which has not been previously studied. This study was also designed to explore novel biomarkers to improve the status of GC, aiming to supplement evidence for identifying novel therapeutic regimen for GC. We present the following article in accordance with the ARRIVE reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-723/rc).
Methods
Clinical patient samples
We selected 10 GC patients who underwent surgery the Sanya Central Hospital of Hainan Province from July 2018 to July 2019. Both GC and paired normal adjacent tissues were collected from each patient. All samples were confirmed as GC by pathologists from the hospital. All samples were rinsed with normal saline and stored in liquid nitrogen at −80 °C. All sample patients did not receive preoperative chemoradiotherapy. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Hainan General Hospital, Hainan Affiliated Hospital of Hainan Medical University (No. 2018-518) and informed consent was taken from all the patients.
Cell culture and treatment
The GC cell lines MGC-803, AGS, and SGC-7901 were purchased from the American Type Culture Collection (ATCC; Rockville, MD, USA). Roswell Park Memorial Institute (RPMI)-1640 complete medium, which contained 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin, was utilized. The cells in the logarithmic growth stage were treated with 0.25% trypsin and then inoculated into 6-well plates. Afterwards, they were incubated at 37 °C with 5% CO2 until the cell confluency rate reached approximately 70%.
Normal gastric fibroblasts (NGFs) and GC-CAF separation: fresh GC tissues were collected (the in vitro time was minimized), fully cleaned with iced phosphate-buffered saline (PBS) three times within 0.5 hours, and separated with a sterile surgical blade. Afterwards, these tissues were placed in 0.1 mg/mL collagenase for 3 hours and trypsinized in a 37 °C incubator (3 h for CAF, 40 min for NGF). After a large number of suspended cell clusters were observed in the trypsinization solution, the cells were filtered with a 200-mesh cell filter and centrifuged at 300 rpm for 5 minutes. Furthermore, the cells were resuspended in the RPMI-1640 complete medium containing 10% FBS and 1% penicillin/streptomycin. Next, the cell density was adjusted to 5×105/mL, inoculated into 6-well plates, and incubated at 37 °C with 5% CO2.
Cell coculture model
To explore the interaction between CAF or NGF with GC cells, we constructed a co-culture model using 0.4 µm transwell chambers. Briefly, GC cells (1×105 cells) were seeded into the lower layer of the culture system (24-well plate), and the CAFs or NGFs (1×105 cells) were seeded into the upper layer of the culture system, namely the transwell chambers. After 24 hours of culturing, the transwell chambers were removed. The GC cells in the lower layer were used for Cell Counting Kit-8 (CCK-8), colony formation assay, and transwell assay, and HMGB3 in the culture medium were detected by enzyme-linked immunosorbent assay (ELISA).
Cell transfection
The isolated NGFs and GC-CAF were cleaned with PBS buffer and trypsinized for two minutes. The NGFs, CAFs, and GC cell lines (MGC-803, AGS, and SGC-7901) were inoculated into 6-well plates at a density of 5×106 cells/well. The CAFs were co-cultured with GC cell lines, and the CAF-coMGC-803 group, the CAF-coAGS group, and the CAF-coSGC-7901 group were obtained. Cell transfection was conducted after cell growth had stabilized. Overexpressed miR-200b (miR-200b mimics) and HMGB3, as well as their corresponding negative controls, were transfected into all the above-mentioned cells according to the Lipofectamine 2000 reagent specification (Thermo Fisher Scientific, Waltham, MA, USA). The Wnt inhibitor (IWR-1, 10 µM, Santa Cruz Biotech., Santa Cruz, CA, USA) was added for experimental purposes. The treated cells were incubated at 37 °C with 5% CO2. Then, cells at 70–80% cell confluency rate were selected using Geneticin (Sigma-Aldrich, St. Louis, MO, USA) for subsequent experiments.
CCK-8 assay
The CCK-8 method was employed to verify the changes in the viability of the stably transfected cells. According to the manufacturer’s regulations, CAFs were incubated in 96-well plates with 2×103 cells/well for 48 hours with 100% humidity and 5% CO2 at 37 °C. A portion of 10 µL CCK-8 solution (Dojindo Molecular Technologies, Kumamoto, Japan) was added to each well. After incubation for 24, 48, 72, and 96 hours, the absorbance value at 450 nm was measured on a spectrophotometer (Bio-Rad Laboratories, Hercules, CA, USA).
Cell colony formation experiment
The stably-transfected CAF-co-cultured cells were inoculated into 6-well plates with 1×103 cells/well. Then, they were cultured in RPMI-1640 complete medium containing 10% FBS and 1% penicillin/streptomycin. After two weeks, the medium was discarded. The cells were washed twice with PBS, immobilized with 4% paraformaldehyde for 10 minutes, and stained with 0.1% crystal violet for 15 minutes. After staining, the stained colonies were imaged and counted with an optical microscope (Olympus, Tokyo, Japan).
Transwell assay
Cell invasiveness was determined by transwell assay. The stably-transfected CAF-co-cultured cells were added to the upper chamber of the transwell pre-coated with Matrigel (LI-COR Biosciences, Lincoln, NE, USA), and the lower chamber received RPMI-1640 complete medium containing 10% PBS. After incubation at 37 °C for 24 hours, the chambers were removed, and the uninvaded cells in the upper chamber were gently wiped off, fixed with methanol for 30 minutes, stained with 0.1% crystal violet solution for 20 minutes, and washed with PBS. The number of invading cells was calculated under a microscope (Olympus).
Western blot
After cell treatment, the medium was discarded. The protein was extracted with the radioimmunoprecipitation assay (RIPA) lysate (Beyotime Biotechnology, Shanghai, China) and quantified by the bicinchoninic acid (BCA) method (Pierce, Appleton, WI, USA). A total of 50 g total protein was sampled in 12% of polyacrylamide gel for 100 V electrophoresis for 2 hours and then transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Bedford, MA, USA). After the membranes had been blocked for 1 hour with 5% skimmed milk at room temperature (RT), they were cleared with tris-buffered saline with Tween 20 (TBST) three times (10 min for each time) and incubated with the primary Anti-E-Cadherin antibody (1:1,000, ab231303, Abcam, Cambridge, MA, USA), Anti-Claudin 1 antibody (1:1,000, ab211737, Abcam), Anti-SLUG antibody (1:1,000, ab27568, Abcam), Anti-Twist antibody (1:1,000, ab49254, Abcam), Anti-p-ERK1/2 antibody (1:1,000, ab201015, Abcam), Anti-ERK1/2 antibody (1:1,000, ab17942, Abcam), Anti-p-JNK antibody (1:1,000, ab4821, Abcam), Anti-JNK antibody (1:1,000, ab124956, Abcam), Anti-TCF4 antibody (1:1,000, ab217668, Abcam), Anti- Fibrillarin antibody (1:1,000, ab4655, Abcam), Anti-c-Myc antibody (1:1,000, ab32072, Abcam), Anti-β-catenin antibody (1:1,000, ab32572, Abcam), Anti-Cyclin D1 antibody (1:1,000, ab16661, Abcam), Anti-HMGB3 antibody (1:1,000, AF1533a, San Diego, USA), and Anti-GAPDH antibody (1:1,000, ab181603, Abcam) at 4 °C overnight. After washing with TBST, the membranes were incubated with Goat Anti-Rabbit IgG H&L (1:3,000, ab150077, Abcam) for 1 hour at RT. Subsequently, the membranes were rinsed three times for 10 minutes each time. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as an internal reference. At last, ImageQuant software (Molecular Dynamics, Sunnyvale, CA, USA) was employed to analyze the gray values of each protein. The uncropped bolts are shown in the Supplementary file (Figures S1-S4).
ELISA
The culture medium of GC cells cocultured with CAFs were harvested, centrifuged to remove cells or debris, then the HMGB3 level in the supernatant was determined by Human HMGB3 ELISA kit (cat# EH0806) (Wuhan FineTest Biotechnology Co., Ltd., Wuhan, China). All procedures were performed according to the manufacturer’s instructions.
Chemosensitivity measurement
The treated cells were inoculated into 96-well plates at 1×104 cells/well. Each group was prepared in triplicate, and 25–200 µg/mL of CDDP was added to the plates. After culturing for 48 hours, the cells were incubated with 5 mg/mL of 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide (MTT) solution (20 µL) for 4 hours. Then, 100 µL of dimethyl sulfoxide (DMSO; Thermo Fisher) was added and incubated for 10 minutes. Absorbance value was examined at 540 nm using a microplate reader (Bio-Rad, USA). The drug sensitivity of the cells was assessed by comparing the half-maximal inhibitory concentration (IC50).
Dual-luciferase reporter assay
The CAF-co-cultured cells were inoculated into 96-well plates at 3×104 cells/well. After culturing for 24 hours, luciferase reporter vectors (HMGB3-MT and HMGB3-WT) and miR-200b mimics were co-transfected into the cells. After transfection for 48 hours, the luciferase activity was examined according to the luciferase activity detection kit instructions (Promega, Madison, WI, USA). The luciferase activity = firefly luciferase activity value/renilla luciferase activity value.
Quantitative reverse transcription polymerase chain reaction
Total cellular RNA was isolated with the TRIzol method (Invitrogen, Carlsbad, CA, USA) and was reversely transcribed into complementary DNA (cDNA) with a TOYOBO reverse transcription kit (TOYOBO, Osaka, Japan). Quantitative real-time polymerase chain reaction (qRT-PCR) was conducted using the SYBR® Premix Ex Taq™ II Kit (Takara, Otsu, Japan). We used GAPDH as the internal reference, and the molecular primers were listed in Table 1.
Table 1
| Name | Primer sequences |
|---|---|
| miR-200b | Forward: 5'-GCGGCTAATACTGCCTGGTAA-3' |
| Reverse: 5'-GTGCAGGGTCCGAGGT-3' | |
| HMGB3 | Forward: 5'-GTCCGCTTATGCCTTCT-3' |
| Reverse: 5'-CATCGTCTTCCACCTCT-3' | |
| Axin2 | Forward: 5'-TGACTCTCCTTCCAGATCCCA-3' |
| Reverse: 5'-TGCCCACACTAGGCTGACA-3' | |
| U6 | Forward: 5'-GGTCGGGCAGGAAAGAGGGC-3' |
| Reverse: 5'-CTAATCTTCTCTGTATCGTTCC-3' | |
| GAPDH | Forward: 5'-AGAAGGCTGGGGCTCATTTG-3' |
| Reverse: 5'-AGGGGCCATCCACAGTCTTC-3-3' |
Tumorigenesis in nude mice
The animal experiments were granted by the Ethics Committee of Hainan General Hospital, Hainan Affiliated Hospital of Hainan Medical University (No. 2018-518), in compliance with national guidelines for the care and use of animals. A protocol was prepared before the study without registration. A total of 20 female BALB/cASlacnt mice (4–6-week-old) were purchased from the Animal Experimental Center of Shandong University (Jinan, China) and raised in specific-pathogen-free (SPF) environments. During the experiment, the mice were provided with adequate food and water. Under aseptic conditions, CAF-coMGC-803, CAF-coAGS, and CAF-coSGC-7901 in the logarithmic growth phase were made into a single-cell suspension with 0.9% normal saline, and the cell concentration was adjusted to 5×107/mL. Then, 0.2 mL of the cell suspension was injected subcutaneously into the right foreleg axilla of each nude mouse with a 1 mL-syringe. Nude mice in the chemotherapy group received 3 mg/kg CDDP (Sigma-Aldrich, St. Louis, MO, USA) orally on days 8, 15, and 22. The general conditions of the nude mice, including spirit, diet, activity, defecation, and urination, were observed within five weeks. The long and short diameters of the tumor were measured with vernier caliper every five days from day 10th after the injection. After five weeks, the nude mice were sacrificed by cervical dislocation. The long and short diameters and weight of the tumor were measured. The tumor volume = long diameter of tumor × short diameter of tumor2/2.
Immunofluorescence
The formed tumor tissues were collected, fixed with 4% formaldehyde for 24 hours, and embedded in paraffin and cut into 4 µm sections. Next, the sections were permeabilized with 0.5% Triton X-100 in PBS for 15 minutes and blocked with 10% normal donkey serum/PBS for 30 minutes at RT. After that, the tissues were incubated with rabbit Anti-ERK1 (phospho T202) + ERK2 (phospho T185) antibody (ab201015) and rabbit Anti-JNK1 + JNK2 + JNK3 (phospho T183+T183+T221) antibody (ab124956) (dilution: 1:100) for 12 hours at 4 °C. Then the sections were washed three times with PBS and incubated with Donkey polyclonal Secondary Antibody to Rabbit IgG - H&L (Alexa Fluor® 488) for 1 hour at RT (ab150073; Abcam, Cambridge, MA, USA). After washing, the nuclei were stained with 4',6-diamidino-2-phenylindole (DAPI). The cells were then observed by a fluorescence microscope (Olympus Optical Co., Ltd., Tokyo, Japan).
Statistical analysis
The statistical Software GraphPad Prism 8 (GraphPad Software Inc., San Diego, CA, USA) was utilized to analyze all the experimental data in this study, and the data were expressed as mean ± standard deviation (). Student’s t-test was adopted to compare the differences between two groups, and one-way ANOVA was used to compare the differences among multiple groups. Tukey’s post-hoc test was employed to analyze the differences between two groups. A P value <0.05 indicated statistical significance.
Results
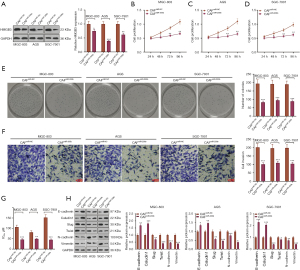
CAFs promoted GC cell growth, EMT, and chemoresistance
The GC-CAFs were co-cultured with MGC-803, AGS, and SGC-7901 cells to study the effect of CAFs on GC cells. The results of CCK-8 showed that CAFs co-cultured with GC cell lines significantly promoted GC cell proliferation (P<0.05, Figure 1A-1C). The results of the colony formation experiment were similar to those of CCK-8. The CAF co-culture significantly increased GC cell viability (P<0.05, Figure 1D). The transwell assay demonstrated that GC cell invasion was significantly enhanced after CAF co-culture (P<0.05, Figure 1E). The MTT assay revealed that CAF co-culture significantly elevated the IC50 of GC cells to CDDP treatment (P<0.05, Figure 1F). DNA RT-PCR data suggested that CAF co-culture promoted the mRNA levels of damage repair-related genes, including RAD51 and BRCA2 in GC cells (all P<0.05, Figure 1G). Western blot confirmed that, compared with the control group, E-cadherin and Caludin1 in CAF-co-cultured-GC cells were down-regulated, while Twist and Slug were up-regulated (all P<0.05, Figure 1H).
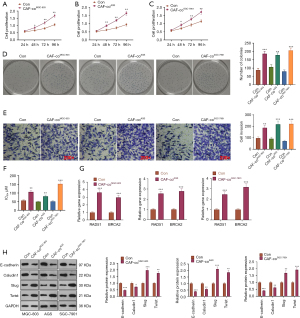
HMGB3 was up-regulated in CAFs co-cultured with GC cells
We employed qRT-PCR, ELISA, and western blot were employed to detect the mRNA and protein levels of HMGB3 in CAFs or the co-culture medium. Interestingly, HMGB3 was significantly overexpressed in CAFs when co-cultured with MGC-803, AGS, and SGC-7901 cells (Figure 2A,2B). In the co-culture medium, the CAFs co-cultured with GC cells released more HMGB3 (Figure 2C). Moreover, HMGB3 was overexpressed in multiple tumors, including stomach adenocarcinoma (STAD) [data from Gene Expression Profiling Interactive Analysis (GEPIA), Figure S1A,S1B]. The higher level of HMGB3 was associated with poorer survival of STAD patients (Figure S1C). The Human Protein Atlas database also showed that HMGB3 was negatively expressed in normal stomach tissues while had a moderate expression in GC tissues (Figure S1D). Moreover, we added the correlation analysis of HMGB3 with immune cells, including B cells, CD8+ T cell, CD4+ T cell, macrophages, neutrophil and dendritic cell via TIMER2.0 (http://timer.comp-genomics.org/) (23). The data showed that HMGB3 has negative relationship with CD8+ T cell (partial correlation=-0.191, P=2.3×10-4) and CD4+ T cell (partial correlation =−0.241, P=3.13×10-6) (Figure S2A). HMGB3 alteration changes both CD8+ T cell and CD4+ T cell infiltration (Figure S2B).
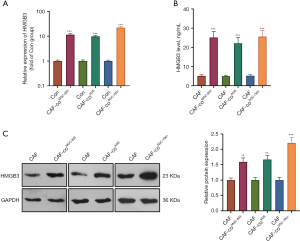
Inhibiting HMGB3 weakened the promoting effects of CAFs on GC cells
We probed the impact of HMGB3 on GC cells co-cultured with CAFs. An HMGB3 knockdown model was established in CAFs (Figure 3A). Next, the CAFs with down-regulated HMGB3 were co-cultured with GC cells, and it was found that HMGB3 was down-regulated in GC cells after the CAF co-culture compared with that of the CAFsi-NC group (P<0.05, Figure 3B,3C). Then, the CCK-8 assay, colony formation experiment, and transwell assay were conducted to determine GC cell proliferation, viability, and invasion, respectively. Interestingly, knocking down HMGB3 restrained the malignant biological behaviors in GC cells co-cultured with CAFs, which was mainly manifested as decreased GC cell proliferation, viability, and invasion (P<0.05, Figure 3D-3H). The MTT assay indicated that knocking down HMGB3 in CAFs attenuated the IC50 of CAF-co-cultured GC cells (P<0.05, Figure 3I). Moreover, western blot revealed that, compared with the CAFsi-NC group, knocking down HMGB3 up-regulated E-cadherin and Caludin1, yet the same action down-regulated Slug, Twist, N-cadherin, and Vimentin (P<0.05, Figure 3J).
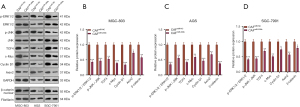
CAFs promoted ERK1/2, JNK, and activation of the Wnt/β-catenin pathway on GC cells
We analyzed the correlations of the HMGB3 level with the expression of ERK1/2 (MAPK1), JNK (MAPK8), β-catenin (CTNNB1), c-Myc (MYC), and Cyclin (CCND1) in STAD through the GEPIA database. Interestingly, HMGB3 had a positive correlation with those five proteins in STAD (Figure S3A-S3E), suggesting that HMGB3 regulated those pathways. Next, western blot was adopted to test the levels of ERK1/2, JNK, and Wnt/β-catenin (including β-catenin, c-Myc, Cyclin, Axin2, and β-catenin) in GC cells. As a result, p-ERK1/2/p-ERK, p-JNK/JNK, TCF4, c-Myc, Cyclin, Axin2, and β-catenin were all significantly up-regulated in the CAF-co-cultured GC cells compared with that of the control group (P<0.05, Figure 4A). In contrast, the profiles of p-ERK1/2/p-ERK, p-JNK/JNK, TCF4, c-Myc, Cyclin, Axin2, and β-catenin were down-regulated in the CAF group with HMGB3 knockdown (P<0.05, Figure 4B). Meanwhile, the Wnt inhibitor (IWR-1) repressed the levels of the above proteins (P<0.05, Figure 4C).
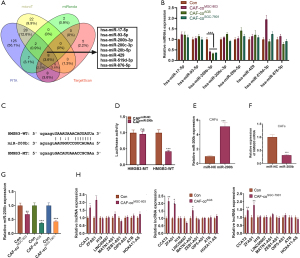
MiR-200b targeted HMGB3 and inhibited its expression
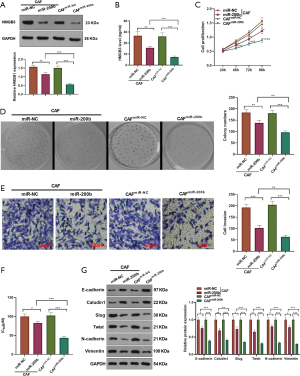
We further investigated the relationship between miR-200b and HMGB3. By querying the StarBase database (https://www.starbase.info/index.html), we found that HMGB3 was a vital downstream target of eight miRNAs, including hsa-miR-17-5p, hsa-miR-93-5p, hsa-miR-200b-3p, hsa-miR-200c-3p, hsa-miR-20b-5p, hsa-miR-429, hsa-miR-519d-3p, and hsa-miR-876-5p (Figure 5A). We performed qRT-PCR to detect those miRNAs in CAFs co-cultured with GC cells. Interestingly, miR-200b-3p was significantly repressed by GC cell co-culturing (Figure 5B). The binding relationship between miR-200b-3p and HMGB3 is shown in Figure 5C. Then, the dual-luciferase reporter assay was used to confirm their targeted relationship. The results showed that miR-200b mimics inhibited the luciferase activity of CAFs transfected with HMGB3-WT but had no inhibitory effect on HMGB3-MT transfected CAFs (P<0.05, Figure 5D). Furthermore, a miR-200b overexpressed CAFs model was constructed (Figure 5E). We found that overexpression of miR-200b significantly reduced HMGB3 expression in CAFs (Figure 5F). Additionally, RT-PCR data showed that miR-200b was downregulated (Figure 5G), whereas lncRNA CCAT2 and lncRNA ZFAS1 (Figure 5H) were significantly upregulated in GC cells when co-cultured with CAFs.
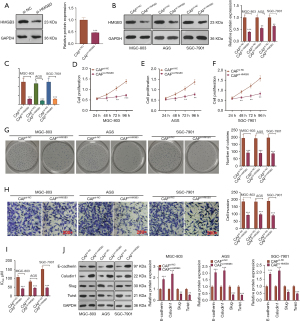
Overexpressing miR-200b attenuated the promotive effects mediated by CAFs on GC
We analyzed the miR-200b level in different STAD patients’ overall survival. It was found that STAD patients with lower levels of miR-200b had poorer survival and shorter survival time (P<0.05, Figure S4). For investing the role of fibroblasts on GC cells, we co-cultured GC cells with normal gastric fibroblasts (NGF). NGF have no significant effects on GC cell proliferation, viability, invasion, CDDP-chemosensitivity, and EMT (Figure S5A-S5H) Furthermore, we probed the role of miR-200b in CAF-co-cultured GC cells. The results of western blot showed that compared with the CAFmiR-NC group, CAFmiR-200b co-cultured GC cells had significantly inhibited HMGB3 expression (P<0.05, Figure 6A). Meanwhile, GC cell proliferation, viability, invasion, CDDP-chemosensitivity, and EMT were analyzed by CCK-8, colony formation, MTT assay, transwell assay, and western blot, respectively. The results illustrated that the transfection of miR-200b mimics in CAFs repressed the CAFs-mediated promotion of GC cell viability and invasion, up-regulated E-cadherin and Caludin1, and down-regulated Slug, Twist, N-cadherin, and Vimentin (P<0.05, Figure 6B-6H).
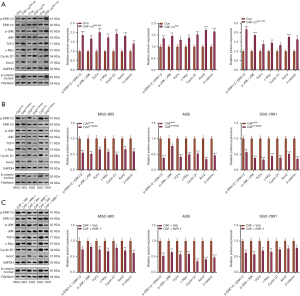
Overexpressing miR-200b inactivated the Wnt pathway in GC
The expression of ERK1/2, JNK, and Wnt/β-catenin was detected by western blot to clarify the specific mechanism of miR-200b’s inhibitory effect on CAFs-GC. The experimental results showed that CAFsmiR-200b significantly reduced the phosphorylated levels of ERK1/2 and JNK, and reduced β-catenin, c-Myc, and Cyclin D1 in GC cells (Figure 7A-7D).
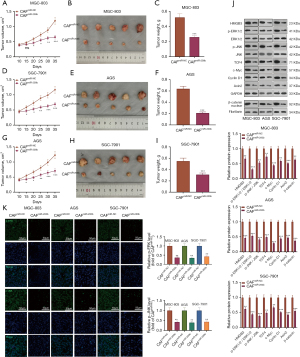
Overexpressing miR-200b abated the oncogenic effects of CAFs in vivo
Finally, we conducted in vivo experiments to verify CAFs-mediated effects in GC cell growth. Three GC cell lines co-cultured with CAFs were made into suspensions and then injected subcutaneously into the axilla of nude mice. Then, the tumor growth was observed and recorded. Interestingly, the growth rate, volume, and weight of tumors in nude mice transfected with miR-200b were lower than those in the miR-NC group (P<0.05, Figure 8A-8I). Additionally, western blot was implemented to detect the expression of the ERK1/2, JNK, and Wnt/β-catenin pathways in tumor tissues. The results showed that transfection with miR-200b mimics significantly inactivated these pathways compared with those of the control group (P<0.05, Figure 8J). Moreover, we measured p-ERK1/2 and p-JNK levels in the formed tumor tissues via immunofluorescence. The results indicated that both p-ERK1/2 and p-JNK were downregulated in the CAFmiR-200b group (compared with the CAFmiR-NC group, P<0.05, Figure 8K).
Overexpressing miR-200b restrained GC progression by inhibiting CAF-released HMGB3
To clarify the inhibitory effect of miR-200b on GC by interfering with HMGB3 in CAFs, the GC cells were transfected with miR-200b (or miR-200b mimics) and co-cultured with CAFs. We conducted ELISA and western blot to examine the HMGB3 expression. As a result, both the transfection of miR-200b into GC cells and the transfection of miR-200b mimics into CAFs reduced HMGB3 expression in GC cells (co-cultured with CAFs) and the release of HMGB3 in the coculture medium (Figure 9A,9B). The CCK-8 assay and cell colony formation experiment showed that the cell viability of the miR-200b+CAF group was significantly weaker than that of the miR-NC group, and that of the CAFmiR-200b group was further weakened (vs. the miR-200b+CAF group) (P<0.05, Figure 9C,9D). The results of transwell assay showed that the number of cell invasion in the miR-200b+CAF group was significantly reduced (P<0.01, vs. miR-NC+CAF group). Similarly, the cell invasion in the CAFmiR-200b group was lower than that in the miR-200b+CAF group (P<0.05, Figure 9E). The MTT assay demonstrated that the IC50 of CDDP in the miR-200b+CAF group was lower than that of the miR-NC+CAF group, and it was lower in the CAFmiR-200b group compared with that of the miR-200b+CAF group (P<0.05, Figure 9F). The results of western blot illustrated that compared with the miR-NC+CAF group, E-cadherin and Caludin1 were up-regulated, while Slug, Twist, N-cadherin, and Vimentin were down-regulated in the CAF+miR-200b group. The above-mentioned proteins in the CAFmiR-200b group showed the same expression trend, and the E-cadherin and Caludin1 proteins were lower, while the expression of Slug, Twist, N-cadherin, and Vimentin was higher than the miR-200b+CAF group (P<0.05, Figure 9G). Moreover, we compared these results with the role of NGF in affecting the malignant behaviors of GC cells. As shown in Figure S4, no significant changes of cell proliferation were seen in GC cells (Figure S5A,S5B), invasion (Figure S5C), sensitivity to CDDP (Figure S5D) and HMGB3 expression (Figure S5E,S5F), and EMT process (Figure S5G,S5H).
Discussion
No specific symptoms are observed in the early stage of GC, and most GC patients have developed to the advanced stage by the time of definite diagnosis. Due to the lack of surgical indications, most patients with advanced GC can only be treated with chemotherapy. Although CDDP is a first-line chemotherapeutic drug in the clinical treatment of advanced GC, long-term CDDP treatment will lead to a decrease in the body’s sensitivity to drugs and further cause tumor recurrence or metastasis, which brings great challenges to the treatment (24). Another risk factor for the occurrence and development of GC is EMT (25). Epithelial cells acquire strong migrative, invasive, and anti-apoptotic capabilities when transiting into mesenchymal cells, so EMT is considered an important target in GC treatment (26,27). During EMT, certain epithelial cell markers, such as E-cadherin, Caludin1, and ZO1, are up-regulated, and N-cadherin, Slug, and Twist are down-regulated. Therefore, this paper focuses on the resistance of miR-200b on GC by targeting HMGB3 to regulate CAFs.
The TEM is the hotbed of tumor growth, which is composed of cancer cells, stromal cells (fibroblasts, immune cells, pro-inflammatory cells, etc.), and non-cellular components such as ECM (28). The fibroblasts activated in tumors are referred to as CAFs, which are the most abundant components of TEM. The CAFs regulate TEM, change tumor metabolism, trigger tumors, deposit various ECM components, stimulate angiogenesis, mediate chemoresistance, and provide a solid support for cancer cell metastasis and invasion (29,30). As confirmed by the study, tumor cells supported by CAFs are more aggressive than those lacking nutrition (31). Li et al. showed that the lysyl oxidase produced by CAFs promotes GC liver metastasis by promoting the interaction between GC cells and CAFs (32). Kurashige et al. suggested that CAFs stimulate gastric cancer invasion and peritoneal dissemination (33). Inhibiting protein synthesis in CAFs improves the chemosensitivity of pancreatic ductal adenocarcinoma to gemcitabine (34). DNA damage repair-related genes, such as RAD51 (35) and BRCA2 (36), play a role in chemoresistance of tumor cells. We isolated CAFs from fresh GC tissues and carried out experiments and found that CAFs promoted GC cell proliferation, invasion, and EMT, increased its chemoresistance to CDDP, and promoted both RAD51 and BRCA2 expression, which was closely related to the role of CAFs in activating the ERK1/2, JNK, and Wnt/β-catenin signaling pathways, suggesting that CAFs are a potentially important target for treating GC.
MiRNAs are involved in various biological mechanisms, including cell differentiation, proliferation, migration, apoptosis, and angiogenesis, among others. It is worth noting that miRNAs contribute to tumor inhibition or promotion, which mainly depends on cancer types or downstream targets (37,38). Many miRNAs have been found to inhibit GC progression. For example, miR-502-5p targets SP1 and inhibits GC cell proliferation, migration, and invasion, thus acting as a tumor suppressor gene (39). For another example, miR-145-5p represses the nod-like-receptor axis by targeting ANGPT2, thus reducing the proliferation, migration, and invasion of GC epithelial cells (40). Interestingly, miRNAs also weaken GC’s chemoresistance to drugs. Zhang et al. found that solasonine inhibits GC cell proliferation and improves its chemosensitivity to paclitaxel by up-regulating miR-486-5p (41). In addition, Fang et al. indicated that overexpressing miR-130a-5p reduces the malignant proliferation and invasion of GC cells, promotes apoptosis, and strengthens the chemosensitivity of GC to CDDP by targeting CCL22 (42). Also, miRNAs mediate EMT in GC. It has been shown that miR-665 inhibits EMT, tumorigenesis, and GC evolvement by targeting CRIM1 (43). One other study has found that up-regulated miR-221-5p targets and inhibits DDR1, thus inhibiting GC cell proliferation, invasion, migration and EMT, and improving its chemosensitivity to CDDP (44). Some researchers have shown that miR-200b is down-regulated in GC, and its abnormal expression impairs GC cell growth and invasion. Therefore, miR-200b is considered a valuable marker for the prognosis of GC (45). Unfortunately, the effect of miR-200b on GC remains elusive. Our experimental results showed that miR-200b was down-regulated in the CAFs isolated from GC. Meanwhile, overexpressing miR-200b targeted and abated HMGB3, thus weakening the CAF-induced GC cell proliferation, invasion, and EMT, reducing tumor growth, improving the sensitivity of GC cells to CDDP, and inactivating the ERK1/2, JNK and Wnt/β-catenin pathways in vitro and in vivo. Moreover, the anti-tumor effect of miR-200b transfection on GC cells via inhibition of HMGB3 in CAF co-cultured GC cells was stronger than that of direct miR-200b intervention on GC cells, indicating that miR-200b contributes to the treatment of GC.
MiRNAs exert negative regulatory effects on genes by binding to the 3'-untranslated region of the downstream target. By querying the database, we found that HMGB3 is an important downstream target of miR-200b. Coincidentally, a recent study has found that up-regulated miR-200b attenuates cell proliferation and migration in human hepatocellular carcinoma by targeting and abating HMGB3 (46). These findings indicate that the miR-200b/HMGB3 axis has the potential to repress malignant tumors. Research has identified HMGB3 as an oncogene, and it has been found that HMGB3 is up-regulated in lung squamous cell carcinoma, and PITPNA-AS1 boosts the proliferation and migration of lung squamous cell carcinoma cells by recruiting TAF15 to stabilize HMGB3 mRNA (47). A recent study has confirmed that HMGB3 is up-regulated in breast cancer, while miR-205 suppresses breast cancer cell proliferation and metastasis by targeting HMGB3 (48). Yamada et al. have claimed that miR-205-5p attenuates the aggressiveness of prostate cancer cells by inhibiting HMGB3 (49), indicating that HMGB3 is an effective target for the treatment of certain tumors. Significantly, another study has shown that knocking down HMGB3 dampens GC cell proliferation and migration, promotes apoptosis, and enhances the sensitivity of GC to CDDP and paclitaxel (50). T cell infiltration in tumor microenvironment greatly affects gastric cancer development by regulating tumor cell growth, EMT, and metastasis (51-53). A previous study has found increased stromal and immune scores and increased infiltration of CD4+ T cell, CD8+ T cell, cancer-associated fibroblast, and macrophage in gastric cancer tissues with high risk score of EMT (54). Interestingly, HMGB3 has negative relationship with CD8+ T cell and CD4+ T cell, and HMGB3 alteration changes both CD8+ T cell and CD4+ T cell infiltration. Our in-vitro experiments confirmed that HMGB3 was overexpressed in CAFs separated from GC cells. Knocking down HMGB3 weakened GC cell proliferation, invasion, and EMT and enhanced its chemosensitivity to CDDP. Moreover, knocking down HMGB3 down-regulated the expression levels of ERK1/2, JNK, p38, and the Wnt downstream proteins including c-Myc, Cyclin D1, and β-catenin, indicating that suppressing HMGB3 is a potential treatment for GC.
Considering the crucial roles of lncRNA in cancer via targeting miRNAs, we have also detected several lncRNAs that target miR-200b, including lncRNA CCAT2 (55), lncRNA ZFAS1 (56), lncRNA H19 (57), lncRNA LINC00667 (58), lncRNA MATN1-AS1 (59), lncRNA ZEB1-AS1 (60), lncRNA OIP5-AS1 (61), lncRNA ATB (62), lncRNA HOXA11-AS (63). Our data showed that lncRNA CCAT2 and lncRNA ZFAS1 were significantly upregulated in CAFs when co-cultured with GC cells. Therefore, it is supposed that lncRNA CCAT2 and lncRNA ZFAS1 potentially promote HMGB3 expression in CAFs by targeting and inhibiting miR-200b.
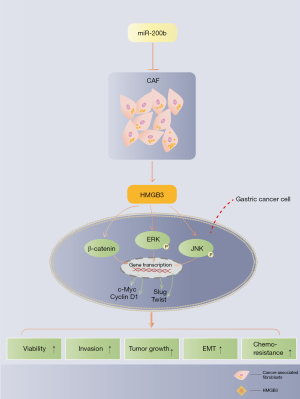
Taken together, this study revealed that miR-200b served as a tumor suppressor gene in GC both in vivo and in vitro. Overexpressed miR-200b reduced HMGB3 release in CAFs. Meanwhile, overexpressing miR-200b attenuated the CAF-induced GC cell proliferation, invasion, and EMT, inhibited tumor growth, and improved the CDDP sensitivity by targeting and negatively regulating HMGB3 (Figure 10). Considering the significance of this study, we believe this miR-200b-HMGB3 axis provides a new reference for developing diagnostic biomarkers of GC, and targeting this axis helps the treatment of GC by reversing tumor cell growth, invasion and chemoresistance.
The major limitation of this study is that the diversity of clinical samples was relatively low. In subsequent studies, the GC clinical samples should be further diversified, aiming to improve the prognosis of GC patients.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-723/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-723/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-723/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Hainan General Hospital, Hainan Affiliated Hospital of Hainan Medical University (No. 2018-518) and informed consent was taken from all the patients. The animal experiments were granted by the Ethics Committee of Hainan General Hospital, Hainan Affiliated Hospital of Hainan Medical University (No. 2018-518), in compliance with national guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Torre LA, Siegel RL, Ward EM, et al. Global Cancer Incidence and Mortality Rates and Trends--An Update. Cancer Epidemiol Biomarkers Prev 2016;25:16-27. [Crossref] [PubMed]
- Thrumurthy SG, Chaudry MA, Chau I, et al. Does surgery have a role in managing incurable gastric cancer? Nat Rev Clin Oncol 2015;12:676-82. [Crossref] [PubMed]
- Yoshida GJ, Azuma A, Miura Y, et al. Activated Fibroblast Program Orchestrates Tumor Initiation and Progression; Molecular Mechanisms and the Associated Therapeutic Strategies. Int J Mol Sci 2019;20:2256. [Crossref] [PubMed]
- LeBleu VS, Kalluri R. A peek into cancer-associated fibroblasts: origins, functions and translational impact. Dis Model Mech 2018;11:dmm029447. [Crossref] [PubMed]
- Zhang Y, Elechalawar CK, Hossen MN, et al. Gold nanoparticles inhibit activation of cancer-associated fibroblasts by disrupting communication from tumor and microenvironmental cells. Bioact Mater 2021;6:326-32. [Crossref] [PubMed]
- Zhang J, Li S, Zhao Y, et al. Cancer-associated fibroblasts promote the migration and invasion of gastric cancer cells via activating IL-17a/JAK2/STAT3 signaling. Ann Transl Med 2020;8:877. [Crossref] [PubMed]
- Dexheimer PJ, Cochella L. MicroRNAs: From Mechanism to Organism. Front Cell Dev Biol 2020;8:409. [Crossref] [PubMed]
- Tsai MM, Wang CS, Tsai CY, et al. Potential Diagnostic, Prognostic and Therapeutic Targets of MicroRNAs in Human Gastric Cancer. Int J Mol Sci 2016;17:945. [Crossref] [PubMed]
- Feng X, Wang Z, Fillmore R, et al. MiR-200, a new star miRNA in human cancer. Cancer Lett 2014;344:166-73. [Crossref] [PubMed]
- Jin HF, Wang JF, Song TT, et al. MiR-200b Inhibits Tumor Growth and Chemoresistance via Targeting p70S6K1 in Lung Cancer. Front Oncol 2020;10:643. [Crossref] [PubMed]
- Amornsupak K, Thongchot S, Thinyakul C, et al. HMGB1 mediates invasion and PD-L1 expression through RAGE-PI3K/AKT signaling pathway in MDA-MB-231 breast cancer cells. BMC Cancer 2022;22:578. [Crossref] [PubMed]
- Zhang P, Lu Y, Gao S. High-mobility group box 2 promoted proliferation of cervical cancer cells by activating AKT signaling pathway. J Cell Biochem 2019;120:17345-53. [Crossref] [PubMed]
- Gong W, Guo Y, Yuan H, et al. HMGB3 is a Potential Therapeutic Target by Affecting the Migration and Proliferation of Colorectal Cancer. Front Cell Dev Biol 2022;10:891482. [Crossref] [PubMed]
- Lin T, Zhang Y, Lin Z, et al. Roles of HMGBs in Prognosis and Immunotherapy: A Pan-Cancer Analysis. Front Genet 2021;12:764245. [Crossref] [PubMed]
- Fang J, Ge X, Xu W, et al. Bioinformatics analysis of the prognosis and biological significance of HMGB1, HMGB2, and HMGB3 in gastric cancer. J Cell Physiol 2020;235:3438-46. [Crossref] [PubMed]
- Nemeth MJ, Curtis DJ, Kirby MR, et al. Hmgb3: an HMG-box family member expressed in primitive hematopoietic cells that inhibits myeloid and B-cell differentiation. Blood 2003;102:1298-306. [Crossref] [PubMed]
- Nemeth MJ, Kirby MR, Bodine DM. Hmgb3 regulates the balance between hematopoietic stem cell self-renewal and differentiation. Proc Natl Acad Sci U S A 2006;103:13783-8. [Crossref] [PubMed]
- Lv Y, Lv M, Ji X, et al. Down-regulated expressed protein HMGB3 inhibits proliferation and migration, promotes apoptosis in the placentas of fetal growth restriction. Int J Biochem Cell Biol 2019;107:69-76. [Crossref] [PubMed]
- Zhang Z, Chang Y, Zhang J, et al. HMGB3 promotes growth and migration in colorectal cancer by regulating WNT/β-catenin pathway. PLoS One 2017;12:e0179741. [Crossref] [PubMed]
- Mukherjee A, Huynh V, Gaines K, et al. Targeting the High-Mobility Group Box 3 Protein Sensitizes Chemoresistant Ovarian Cancer Cells to Cisplatin. Cancer Res 2019;79:3185-91. [Crossref] [PubMed]
- Tang HR, Luo XQ, Xu G, et al. High mobility group-box 3 overexpression is associated with poor prognosis of resected gastric adenocarcinoma. World J Gastroenterol 2012;18:7319-26. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Wagner AD, Syn NL, Moehler M, et al. Chemotherapy for advanced gastric cancer. Cochrane Database Syst Rev 2017;8:CD004064. [PubMed]
- Wang H, Zhang B, Wang X, et al. TRPV4 Overexpression Promotes Metastasis Through Epithelial-Mesenchymal Transition in Gastric Cancer and Correlates with Poor Prognosis. Onco Targets Ther 2020;13:8383-94. [Crossref] [PubMed]
- Pastushenko I, Blanpain C. EMT Transition States during Tumor Progression and Metastasis. Trends Cell Biol 2019;29:212-26. [Crossref] [PubMed]
- Liao TT, Yang MH. Revisiting epithelial-mesenchymal transition in cancer metastasis: the connection between epithelial plasticity and stemness. Mol Oncol 2017;11:792-804. [Crossref] [PubMed]
- Lorusso G, Rüegg C. The tumor microenvironment and its contribution to tumor evolution toward metastasis. Histochem Cell Biol 2008;130:1091-103. [Crossref] [PubMed]
- Öhlund D, Elyada E, Tuveson D. Fibroblast heterogeneity in the cancer wound. J Exp Med 2014;211:1503-23. [Crossref] [PubMed]
- Yoshida GJ. Regulation of heterogeneous cancer-associated fibroblasts: the molecular pathology of activated signaling pathways. J Exp Clin Cancer Res 2020;39:112. [Crossref] [PubMed]
- Curtis M, Kenny HA, Ashcroft B, et al. Fibroblasts Mobilize Tumor Cell Glycogen to Promote Proliferation and Metastasis. Cell Metab 2019;29:141-155.e9. [Crossref] [PubMed]
- Li Q, Zhu CC, Ni B, et al. Lysyl oxidase promotes liver metastasis of gastric cancer via facilitating the reciprocal interactions between tumor cells and cancer associated fibroblasts. EBioMedicine 2019;49:157-71. [Crossref] [PubMed]
- Kurashige J, Mima K, Sawada G, et al. Epigenetic modulation and repression of miR-200b by cancer-associated fibroblasts contribute to cancer invasion and peritoneal dissemination in gastric cancer. Carcinogenesis 2015;36:133-41. [Crossref] [PubMed]
- Duluc C, Moatassim-Billah S, Chalabi-Dchar M, et al. Pharmacological targeting of the protein synthesis mTOR/4E-BP1 pathway in cancer-associated fibroblasts abrogates pancreatic tumour chemoresistance. EMBO Mol Med 2015;7:735-53. [Crossref] [PubMed]
- Zhang B, Niu H, Cai Q, et al. Roscovitine and Trichostatin A promote DNA damage repair during porcine oocyte maturation. Reprod Fertil Dev 2019;31:473-81. [Crossref] [PubMed]
- Zhang Y, Wu H, Yang F, et al. Prognostic Value of the Expression of DNA Repair-Related Biomarkers Mediated by Alcohol in Gastric Cancer Patients. Am J Pathol 2018;188:367-77. [Crossref] [PubMed]
- Balachandran AA, Larcher LM, Chen S, et al. Therapeutically Significant MicroRNAs in Primary and Metastatic Brain Malignancies. Cancers (Basel) 2020;12:2534. [Crossref] [PubMed]
- Chauhan N, Dhasmana A, Jaggi M, et al. miR-205: A Potential Biomedicine for Cancer Therapy. Cells 2020;9:1957. [Crossref] [PubMed]
- Peng X, Wu M, Liu W, et al. miR-502-5p inhibits the proliferation, migration and invasion of gastric cancer cells by targeting SP1. Oncol Lett 2020;20:2757-62. [Crossref] [PubMed]
- Zhou K, Song B, Wei M, et al. MiR-145-5p suppresses the proliferation, migration and invasion of gastric cancer epithelial cells via the ANGPT2/NOD_LIKE_RECEPTOR axis. Cancer Cell Int 2020;20:416. [Crossref] [PubMed]
- Zhang Y, Han G, Cao Y, et al. Solasonine inhibits gastric cancer proliferation and enhances chemosensitivity through microRNA-486-5p. Am J Transl Res 2020;12:3522-30. [PubMed]
- Fang QL, Li KC, Wang L, et al. Targeted Inhibition of CCL22 by miR-130a-5p Can Enhance the Sensitivity of Cisplatin-Resistant Gastric Cancer Cells to Chemotherapy. Cancer Manag Res 2020;12:3865-75. [Crossref] [PubMed]
- Wu KZ, Zhang CD, Zhang C, et al. miR-665 Suppresses the Epithelial-Mesenchymal Transition and Progression of Gastric Cancer by Targeting CRIM1. Cancer Manag Res 2020;12:3489-501. [Crossref] [PubMed]
- Jiang X, Jiang M, Guo S, et al. Promotion of miR-221-5p on the Sensitivity of Gastric Cancer Cells to Cisplatin and Its Effects on Cell Proliferation and Apoptosis by Regulating DDR1. Onco Targets Ther 2020;13:2333-45. [Crossref] [PubMed]
- Tang H, Deng M, Tang Y, et al. miR-200b and miR-200c as prognostic factors and mediators of gastric cancer cell progression. Clin Cancer Res 2013;19:5602-12. [Crossref] [PubMed]
- Wang LK, Xie XN, Song XH, et al. Upregulation of miR-200b Inhibits Hepatocellular Carcinoma Cell Proliferation and Migration by Targeting HMGB3 Protein. Technol Cancer Res Treat 2018;17:1533033818806475. [Crossref] [PubMed]
- Ren P, Xing L, Hong X, et al. LncRNA PITPNA-AS1 boosts the proliferation and migration of lung squamous cell carcinoma cells by recruiting TAF15 to stabilize HMGB3 mRNA. Cancer Med 2020;9:7706-16. [Crossref] [PubMed]
- Elgamal OA, Park JK, Gusev Y, et al. Tumor suppressive function of mir-205 in breast cancer is linked to HMGB3 regulation. PLoS One 2013;8:e76402. [Crossref] [PubMed]
- Yamada Y, Nishikawa R, Kato M, et al. Regulation of HMGB3 by antitumor miR-205-5p inhibits cancer cell aggressiveness and is involved in prostate cancer pathogenesis. J Hum Genet 2018;63:195-205. [Crossref] [PubMed]
- Guo S, Wang Y, Gao Y, et al. Knockdown of High Mobility Group-Box 3 (HMGB3) Expression Inhibits Proliferation, Reduces Migration, and Affects Chemosensitivity in Gastric Cancer Cells. Med Sci Monit 2016;22:3951-60. [Crossref] [PubMed]
- Ntala C, Salji M, Salmond J, et al. Analysis of Prostate Cancer Tumor Microenvironment Identifies Reduced Stromal CD4 Effector T-cell Infiltration in Tumors with Pelvic Nodal Metastasis. Eur Urol Open Sci 2021;29:19-29. [Crossref] [PubMed]
- Gauttier V, Pengam S, Durand J, et al. Selective SIRPα blockade reverses tumor T cell exclusion and overcomes cancer immunotherapy resistance. J Clin Invest 2020;130:6109-23. [Crossref] [PubMed]
- Tian Y, Li Y, Shao Y, et al. Gene modification strategies for next-generation CAR T cells against solid cancers. J Hematol Oncol 2020;13:54. [Crossref] [PubMed]
- Xu H, Wan H, Zhu M, et al. Discovery and Validation of an Epithelial-Mesenchymal Transition-Based Signature in Gastric Cancer by Genomics and Prognosis Analysis. Biomed Res Int 2021;2021:9026918. [Crossref] [PubMed]
- Wu X, Fan Y, Liu Y, et al. Long Non-Coding RNA CCAT2 Promotes the Development of Esophageal Squamous Cell Carcinoma by Inhibiting miR-200b to Upregulate the IGF2BP2/TK1 Axis. Front Oncol 2021;11:680642. [Crossref] [PubMed]
- O'Brien SJ, Fiechter C, Burton J, et al. Long non-coding RNA ZFAS1 is a major regulator of epithelial-mesenchymal transition through miR-200/ZEB1/E-cadherin, vimentin signaling in colon adenocarcinoma. Cell Death Discov 2021;7:61. [Crossref] [PubMed]
- Huang Z, Chu L, Liang J, et al. H19 Promotes HCC Bone Metastasis Through Reducing Osteoprotegerin Expression in a Protein Phosphatase 1 Catalytic Subunit Alpha/p38 Mitogen-Activated Protein Kinase-Dependent Manner and Sponging microRNA 200b-3p. Hepatology 2021;74:214-32. [Crossref] [PubMed]
- Liu P, Chen S, Huang Y, et al. LINC00667 promotes Wilms' tumor metastasis and stemness by sponging miR-200b/c/429 family to regulate IKK-β. Cell Biol Int 2020;44:1382-93. [Crossref] [PubMed]
- Zhu J, Gu W, Yu C. MATN1-AS1 promotes glioma progression by functioning as ceRNA of miR-200b/c/429 to regulate CHD1 expression. Cell Prolif 2020;53:e12700. [Crossref] [PubMed]
- Gao R, Zhang N, Yang J, et al. Long non-coding RNA ZEB1-AS1 regulates miR-200b/FSCN1 signaling and enhances migration and invasion induced by TGF-β1 in bladder cancer cells. J Exp Clin Cancer Res 2019;38:111. [Crossref] [PubMed]
- Kun-Peng Z, Chun-Lin Z, Xiao-Long M, et al. Fibronectin-1 modulated by the long noncoding RNA OIP5-AS1/miR-200b-3p axis contributes to doxorubicin resistance of osteosarcoma cells. J Cell Physiol 2019;234:6927-39. [Crossref] [PubMed]
- Li Z, Wu X, Gu L, et al. Long non-coding RNA ATB promotes malignancy of esophageal squamous cell carcinoma by regulating miR-200b/Kindlin-2 axis. Cell Death Dis 2017;8:e2888. [Crossref] [PubMed]
- Chen JH, Zhou LY, Xu S, et al. Overexpression of lncRNA HOXA11-AS promotes cell epithelial-mesenchymal transition by repressing miR-200b in non-small cell lung cancer. Cancer Cell Int 2017;17:64. [Crossref] [PubMed]
(English Language Editor: J. Jones)


