
Decitabine enhances the tumoricidal potential of TRAIL via the epigenetic regulation of death receptor 4 in gastric cancer
Introduction
Gastric cancer (GC) is one of the most common digestive system malignancies worldwide, and the prognosis of advanced GC patients is often poor. About 49% of all GC-related mortalities occur in East Asia (1,2). Recently, the incidence of GC has begun to decline; however, the overall survival (OS) of GC patients remains low (3,4). To improve the prognosis of GC patients, a large amount of studies are currently examining how to improve the efficacy of drug therapies.
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL, also known as the APO2 ligand, APO2L) is a cytokine of the tumor necrosis factor (TNF) superfamily (5). TRAIL holds promise for cancer therapy, as it has a unique capacity to selectively kill tumor cells, and it has little cytotoxicity on normal cells (6). TRAIL fulfills its function by binding to death receptors (DRs), including DR4 (TRAIL-R1) and DR5 (TRAIL-R2) (7). It kills tumor cells by inducing caspase-dependent apoptosis. However, a number of clinical studies have shown that some solid tumors, including GC, are resistant to TRAIL (6,8). To date, some studies have focused on how to improve the tumor-killing effect of TRAIL and found that combination therapy is a valid method to increase the anti-tumor effect of TRAIL (9).
Decitabine (5-aza-2’-deoxycytidine, DAC) is a natural 2'-deoxycytidine analog of adenosine, which is the strongest known deoxyribonucleic acid (DNA) methylation-specific inhibitor. It can inhibit DNA methyltransferase, modulate gene expression, and affect drug sensitivity (10). Studies have shown that decitabine and cisplatin act synergistically to exert an anti-tumor effect on GC cells by inducing SRY-box-containing gene 2 (Sox2) DNA demethylation (11). Notably, decitabine has been shown to upregulate endogenous TRAIL and enhance TRAIL-induced apoptosis in models of stepwise tumorigenesis (12). Fan et al. showed that the combined use of decitabine and TRAIL inhibits the proliferation of hepatocellular carcinoma (13). However, it is not yet known whether decitabine increases the sensitivity of GC cells to TRAIL.
In this study, we examined the synergistic anti-tumor effect of decitabine and TRAIL in GC. We analyzed the effects of decitabine and TRAIL on tumor cell growth and apoptosis, and preliminarily explored the potential mechanism. We present the following article in accordance with the MDAR reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-928/rc).
Methods
Reagent
DAC was purchased from Selleck (Houston, USA). Soluble recombinant human TRAIL was purchased from PeproTech (London, UK). DR4 and non-targeting scramble small inhibitory ribonucleic acids (siRNAs) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Allophycocyanin (APC)-conjugated anti-DR4 and anti-DR5 antibody, and the isotype control, which were used for the flow cytometry, were purchased from BioLegend (San Diego, CA). The following antibodies were used for western blot: anti-DR4 (Abcam Cat# ab8414, RRID:AB_306549), anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Abcam Cat# ab128915, RRID:AB_11143050), and anti-DR5 (Sigma-Aldrich Cat# HPA023625). The other antibodies used included caspase-7 (Cell Signaling Technology Cat# 9492, RRID:AB_2228313), cleaved-caspase-7 (Cell Signaling Technology Cat# 8438, RRID:AB_11178377), caspase-8 (Cell Signaling Technology Cat# 9746, RRID:AB_2275120), caspase-9 (Cell Signaling Technology Cat# 9502, RRID:AB_2068621), cleaved-caspase-9 (Cell Signaling Technology Cat# 7237, RRID:AB_10895832), PARP (Cell Signaling Technology Cat# 9542, RRID:AB_2160739), and cleaved-PARP (Cell Signaling Technology Cat# 5625, RRID:AB_10699459).
Cell viability assays
Human GC cell lines AGS, SNU-1, SNU-16 and NCI-N87 were obtained from American type culture collection (ATCC). SGC-7901, BGC-823, and MGC-803 were obtained from Cell Research Institute (Shanghai, China). MKN28 and NUGC3 were obtained from Health Science Research Resources Bank (Tokyo, Japan). All the cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) or Roswell Park Memorial Institute medium 1640 (GIBCO BRL, Carlsbad, CA) supplemented with 10% fetal bovine serum and 100 units/mL of penicillin in 5% carbon dioxide at 37 ℃.
To monitor the growth inhibition effect, the IncuCyte ZOOM Live-Cell Analysis System (Essen Bioscience) was used in this study. The cells were pre-incubated with decitabine (2 or 5 µM) or DMEM medium for 48 h, and then seeded into 96-well plates. Next, 20 h later, the cells were incubated with TRAIL (100 ng/mL) and/or decitabine (2 or 5 µM) for 24 h. The IncuCyte System took cell photographs every 2 h, and the cell proliferation curve was depicted by IncuCyte ZOOM software (Essen Bioscience).
To examine the synergistic effects, cell viability was measured using Cell Counting Kit-8 (CCK-8) assays (Dojindo Molecular Technologies, Japan). The GC cells were pre-treated with decitabine and seeded into the 96-well plates mentioned above. Next, the cells were treated with TRAIL (100 or 200 ng/mL) and/or decitabine (2 or 5 µM) for 24 h, and CCK-8 solution was used to measure the absorbance. The synergetic effect of decitabine and TRAIL was evaluated in accordance with Jin’s formula, which states: Q = Ea + b/[Ea + (1 – Ea) × Eb], where Ea, Eb and Ea + b represent the response rate of TRAIL, decitabine, and the combination treatment, respectively (14,15). In this method, Q<0.85, 0.85≤Q<1.15 and Q≥1.15 indicated antagonism, additive effects, and synergism, respectively.
Flow cytometry
Apoptosis was detected using an Annexin V/Propidium Iodide (PI) double staining kit (Dojindo Molecular Technologies, Japan) and analyzed by flow cytometry (BD Accuri C6, USA). The cells were pre-cultured with free medium or free medium supplemented with decitabine (5 µM) for 48 h. An equal quantity of cells were treated with a vehicle, decitabine (5 µM), TRAIL (100 ng/mL), and combination regimens for 24 h. Next, the cells were stained in accordance with the manufacturer’s protocol.
To analyze the effect of decitabine on the surface expression of the DRs, the cells were treated with decitabine or the vehicle for 48 h and then incubated with APC-conjugated antibody for 30 min at 4 ℃. The DRs expression was measured by flow cytometry.
RNA interference
The cells (1×105) were seeded into 6-well plates. The siRNAs (100 nmol/L) were diluted in Opti-MEM™ medium (GIBCO BRL, Carlsbad, CA) mixed with Lipofectamine 2000 reagent (Invitrogen, Carlsbad, USA). After incubation for 6 h, fresh complete medium was replaced for an additional 20 h of cell culture. The transfection effect was confirmed by western blot.
DNA extraction and methylation-specific PCR (MSP)
Genomic DNA was extracted from the GC cells in accordance with the standard protocol for the TIANamp genomic DNA kit (Tiangen, Beijing, China), and the DNA concentration was measured using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, Delavare, USA). Approximately 1 µg of DNA was bisulfite-modified with an EZ DNA methylation-gold kit (Zymo Research Corporaion, Irvine, CA, USA). The bisulfite-treated DNA was analyzed by polymerase chain reaction (PCR) assays. The PCR assays were performed using the primer and PCR conditions previously described (16). The sequences were as follows: DR4-M (sense) 5'-TTCGAATTTCGGGAGCGTAGC-3' and (antisense) 5'-GTAATTCAATCCTCCCCGCGA-3'; DR4-U (sense) 5'-GTAGTGATTTTGAATTTTGGGAGTGTAGT-3' and (antisense) 5'-CTCATAATTCAATCCCCACAA-3'. The PCR products were electrophoresed in agarose gel and visualized under ultraviolet illumination.
Quantitative RT-PCR
Total RNA was isolated using Trizol reagent (Life Technologies, USA), and complementary DNA was synthesized using the SuperScript III reverse transcription kit (Invitrogen, Carlsbad, CA, USA). Quantitative PCR was performed with an ABI7500 fast real-time PCR system (Applied Biosystems, USA) using the SYBRTM Green PCR master mix kit (Applied Biosystems, USA). The primer sequences were as follows (17): DR4 (sense) 5'-CGATGTGGTCAGAGCTGGTACAGC-3' and (antisense) 5'-GGACACGGCAGAGCCTGTGCCATC-3'; DR5 (sense) 5'-GGGAGCCGCTCATGAGGAAGTTGG-3' and (antisense) 5'-GGCAAGTCTCTCTCCCAGCGTCTC-3' and GAPDH (sense) 5'- GAAGGTGAAGGTCGGAGT-3' and (antisense) 5'- GAAGATGGTGATGGGATTTC-3'. The data were normalized to GAPDH and the relative quantitation of gene expression was measured using the comparative cycle threshold (ΔΔCT).
Western blot
The prepared cells were treated with radioimmunoprecipitation assay lysis buffer (Solarbio, China). The proteins were quantified using the bicinchoninic acid protein assay kit (Pierce, Rockford, USA), and stored at –80 ℃ awaiting use. An equivalent amount of protein (20 µg) was loaded to 10–12% polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene fluoride membranes (Millipore, Billerica, MA). The membranes were probed with each primary antibody overnight at 4 ℃, followed by incubation with horseradish peroxidase–conjugated secondary antibodies. Signals were detected using chemiluminescent agents (Pierce, Rockford, USA).
Statistical analysis
The results of the 3 independent experiments are presented as the mean ± standard deviation. the difference between the 2 groups was determined using a two-sided Student’s t-test. P value <0.05 was considered statistically significant. All the statistical analyses were performed using GraphPad Prism software (9.0 version).
Results
Decitabine and TRAIL inhibited cell growth synergistically
To evaluate the synergistic effect of decitabine and TRAIL, AGS and SGC-7901 were employed as TRAIL-resistant GC cell lines based on our previous study (18). The cells were stimulated with various concentrations of decitabine (2 or 5 µM) and TRAIL (100 ng/mL) for the indicated times. The cell growth conditions were photographed (see Figure 1A). Decitabine exhibited a slight growth inhibiting effect on the GC cells. The cell growth was mildly inhibited by TRAIL alone, but effectively reduced in the combination group (see Figure 1B). The CCK-8 assays further confirmed this synergism. The results revealed that decitabine facilitated TRAIL-induced cytotoxicity in a dose-dependent manner (see Figure 1C). According to Jin’s formula, which was used to count the synergetic effect of the 2 drugs (15), the Q values were 2.04 for AGS and 1.31 for SGC-7901 at the combination of 5 µM of decitabine with 100 ng/mL of TRAIL, which had a stronger effect than other dose combination and was chosen for the following experiments.
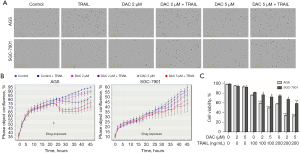
Decitabine sensitized TRAIL-induced apoptosis by activating the caspase-dependent apoptotic pathway
To ascertain whether cell death was due to apoptosis, flow cytometry was performed. The apoptotic rate was significantly increased in the combination treatment group compared to the TRAIL alone group (see Figure 2A,2B). Taking the AGS cell line as an example, the mean apoptotic rate was only 23.06%±0.25% in the TRAIL treatment group, but was 41.97%±1.79% in the combination treatment group. To further confirm this finding and evaluate the apoptosis signals, the expression of the full-length and cleaved-caspase proteins were detected by western blot. As Figure 2C shows, TRAIL mildly activated Caspase-7, -8, -9 and PARP in the GC cells, and the above phenomenon was significantly enhanced after combined treatment, suggesting that decitabine promotes TRAIL-induced apoptosis depending on caspase activation (see Figure 2C).

Decitabine regulates TRAIL sensitivity of GC cells by upregulating DR4
Next, the possible mechanism of decitabine sensitization to TRAIL was investigated. Given that TRAIL acts by binding to DRs, we analyzed the effect of decitabine on TRAIL receptor expression. After the cells were treated with decitabine for 48 h, the expression of DR4, not DR5, increased at the transcriptional and post-transcriptional levels (see Figure 3A,3B). Additionally, DR expression on the cell membrane surface were measured by flow cytometry. The cell surface expression of DR4 was elevated after decitabine exposure (see Figure 3C). Notably, the silencing of DR4 by siRNA reduced the TRAIL-mediated apoptosis and the synergistic effect, indicating that the modulation of DR4 expression takes part in the combination effect of decitabine and TRAIL (see Figure 3D,3E).
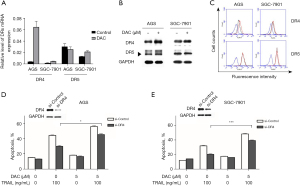
DR4 methylation regulation
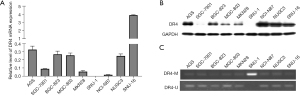
As decitabine is a demethylation drug, we examined the relationship between the DNA methylation level of DR4 and gene expression. The qRT-PCR and western blot were performed to detect the expression of DR4 in the 9 GC cell lines. DR4 was ubiquitously detected in all the GC cells expect SNU-1 (see Figure 4A,4B). The methylation of DR4 gene promoters was detected using methylation-specific PCR. DR4 methylation was detected in SNU-1 cells, which was negatively correlated with its gene expression level (see Figure 4C). As we proved that demethylation regulation enhanced the expression of DR4, these results provide further evidence that decitabine promotes the killing effect of TRAIL on GC through demethylation.
Discussion
TRAIL is an attractive anti-tumor agent. However, drug resistance is a major obstacle to TRAIL-based therapy. GC is a highly heterogeneous solid tumor that is resistant to many drugs. Our previous study showed that about half of the GC cell lines were insensitive to TRAIL-mediated apoptosis signals (18). Multiple resistant mechanisms have been identified. The most common cause of TRAIL resistance is dysfunction of death receptors (19). The defect in caspase protein (20), and overexpression of pro-survival proteins (21) are also linked to TRAIL resistance. In addition, cell survival signals such as transcription factor NF-κB played pivotal role in regulation of TRAIL sensitivity (22). Improving the killing effect of TRAIL on tumor cells is vital in the treatment of GC. Fortunately, combination therapies may be able to be used to reduce the drug resistance of TRAIL. Decitabine is a potent DNA methylation inhibitor that can inhibit tumor cell proliferation and weaken drug resistance by reducing methylation. In this study, we showed that decitabine upregulated DR4 by promoting demethylation and increasing the sensitivity of GC cells to TRAIL.
Combination therapy is an effective way to increase the sensitivity of TRAIL. Previous studies have shown that combined therapy enhances the tumor-killing effect of TRAIL by upregulating apoptosis pathway proteins, inhibiting anti-apoptotic proteins, and regulating phosphorylation pathways. A previous GC study showed that cisatracurium besilate enhanced TRAIL-induced GC apoptosis through tumor protein 53 signaling (23). Li et al. found that 5-fluorouracil (5-FU) enhanced the chemosensitivity of GC to TRAIL by inhibiting the mitogen-activated protein kinase (MAPK) pathway (24). Periplocin upregulated DRs by activating the extracellular signal-regulated protein kinases1/2 pathway (25). Xie et al. combined TRAIL and liquiritin to synergize human GC cells and xenografts in nude mice by enhancing apoptosis and reactive oxygen species generation (26).
Our study showed that decitabine single-drug exposure had a weak inhibitory effect on the growth of GC cells, but when used in combination with TRAIL, it significantly inhibited the growth of tumor cells. This growth inhibitory effect was dose-dependent, and increased as decitabine concentrations increased. Conversely, increasing the concentrations of TRAIL did not enhance cell inhibition. Thus, decitabine exposure changed the molecular characteristics of GC cells, making them more sensitive to the killing effect of TRAIL.
Further apoptosis-related experiments revealed that decitabine enhanced the TRAIL-mediated apoptosis of GC cells, indicating that the growth inhibitory effect was mainly produced by activating the apoptotic pathway. Apoptosis involves multiple aspects, such as pathway gene expression and regulation, apoptosis signal transduction, and the interaction of apoptosis-related molecules. In our study, we found that decitabine combined with TRAIL enhanced the activation of caspase proteins. In terms of pathway gene expression, we found that decitabine upregulated the apoptotic receptor DR4. We also examined the effect of decitabine pretreatment on other apoptotic proteins, including the B-cell lymphoma-2 (Bcl-2) proteins and inhibitor of apoptosis proteins (IAPs); however, no changes in the expression of any related proteins were detected. Overall, decitabine mainly appears to enhance the tumor-killing effect of TRAIL through strengthened caspase-dependent apoptosis signaling by upregulating the TRAIL receptor DR4.
DRs initiate TRAIL apoptotic signals and are important regulators of drug sensitivity. The loss of DR expression by mutation and methylation is one of the main causes of drug resistance. Recently, some studies have shown that drugs can enhance TRAIL sensitivity by upregulating the expression and redistribution of DRs. You et al. found that trichosanthin sensitizes non-small cell lung cancer cells to TRAIL by modulating DR expression and subcellular localization (27). Han et al. verified that the natural compound Periplocin upregulates the expression of DRs in vitro and in vivo, resulting in a high apoptosis rate of esophageal squamous cell carcinoma cells (28).
DR4 and DR5, as 2 receptors for TRAIL, both mediate intracellular stress-induced apoptosis. According to our previous study, DR4 and DR5 have unequal apoptotic signal transduction roles in GC, and DR4 is more positively correlated with TRAIL sensitivity (17). The upregulation of DR4 is important to improve TRAIL resistance in GC. We found that decitabine upregulates the expression of DR4 on cell membranes, which is of great significance in ameliorating the TRAIL resistance of GC.
Epigenetics plays an important role in the process of tumorigenesis and development (29). The abnormal hypermethylation of specific gene is one of the characteristic changes in tumor cells. The hypermethylation of tumor suppressor gene will lead to gene inactivation and molecular changes. At present, DNA methyltransferase inhibitors have led to great advances in the treatment of hematological diseases, such as decitabine, which has been approved by the Food and Drug Administration for the treatment of myelodysplastic syndrome and acute leukemia and achieved satisfactory therapeutic effects (30,31). Small doses of decitabine can maximize the therapeutic effect, and the most important finding is that it can effectively remove DNA methylation and re-express many inactive genes. Under the catalysis of methyltransferase, the transformation of S-adenosine-L-Methionine is a methyl donor, which transfers the methyl group to the 5-carbon atom of cytosine to generate 5-methylcytosine, thereby regulating gene expression (32). Besides, studies have shown that decitabine also played an important role in innate immune response. Decitabine may act as a nucleic acid drug, which has similar functions with anti-virus drugs. The research of Xiao J et al. confirmed that decitabine showed potential antiviral activity by up-regulating the innate antiviral immune response (33).
DNA methylation is one of the most common regulatory mechanisms involved in epigenetic modifications. DNA methylation occurs mainly at the CpG islands. About 70% of the promoters in the human genome are rich in CpG islands. About 60–90% of the CpG islands in the normal genome are methylated (34). Gastric carcinogenesis is associated with multiple epigenomic mutations. Understanding the epigenetic features in GC patients may help us to better treating this disease (35).
DR4 expression is regulated by promoter methylation. The hypermethylation of DR4 gene leads to down-expression of DR4. According to our previous study, DR4 expression level is positively correlated with TRAIL sensitivity. Other studies also proved that hypermethylation of the DR4 gene promoter was responsible for TRAIL resistance (36). In this research, we examined the methylation in the promoter region of the DR4 gene and found that different degrees of methylation levels existed in the GC cell lines. Among these, the SNU-1 cells were most regulated by methylation, and its DR4 expression was completely silenced. The confirmation of the methylation status of DR4 provided further evidence that decitabine regulates DR4 expression through demethylation.
Our study had some limitations. We confirmed that decitabine improves the killing effects of TRAIL on GC cells; however, animal experiments should be carried out to further observe the synergistic effects of the 2 drugs. We will also continue to examine the regulation of the upstream pathway. In the future, the safety and effectiveness of TRAIL-associated compounds might be explored in clinical trials.
Our study confirmed that decitabine can be used as a sensitizing agent of TRAIL by upregulating DR4. These findings may provide novel ways to overcome the drug resistance of TRAIL in treating tumors in clinical practice and may lead to novel ideas for the treatment of GC.
Acknowledgments
Funding: This work is supported by the National Nature Science Foundation of China (Nos. 81902985 to Lin Li and 81402308 to Biao Fan and Lin Li).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-928/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-928/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-928/coif). LL reports grants from national nature science funding (Nos.81902985 and 81402308). BF reports grant from national nature science funding (Nos. 81402308). The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Wang B, He F, Hu Y, et al. Cancer incidence and mortality and risk factors in member countries of the " Belt and Road " initiative. BMC Cancer 2022;22:582. [Crossref] [PubMed]
- Karimi P, Islami F, Anandasabapathy S, et al. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev 2014;23:700-13. [Crossref] [PubMed]
- Li Y, Feng A, Zheng S, et al. Recent Estimates and Predictions of 5-Year Survival in Patients with Gastric Cancer: A Model-Based Period Analysis. Cancer Control 2022;29:10732748221099227. [Crossref] [PubMed]
- Wiley SR, Schooley K, Smolak PJ, et al. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity 1995;3:673-82. [Crossref] [PubMed]
- Dimberg LY, Anderson CK, Camidge R, et al. On the TRAIL to successful cancer therapy? Predicting and counteracting resistance against TRAIL-based therapeutics. Oncogene 2013;32:1341-50. [Crossref] [PubMed]
- Sheridan JP, Marsters SA, Pitti RM, et al. Control of TRAIL-induced apoptosis by a family of signaling and decoy receptors. Science 1997;277:818-21. [Crossref] [PubMed]
- Deng D, Shah K. TRAIL of Hope Meeting Resistance in Cancer. Trends Cancer 2020;6:989-1001. [Crossref] [PubMed]
- Trivedi R, Mishra DP. Trailing TRAIL Resistance: Novel Targets for TRAIL Sensitization in Cancer Cells. Front Oncol 2015;5:69. [Crossref] [PubMed]
- Jabbour E, Issa JP, Garcia-Manero G, et al. Evolution of decitabine development: accomplishments, ongoing investigations, and future strategies. Cancer 2008;112:2341-51. [Crossref] [PubMed]
- Zhu Z, Lin S, Wu X, et al. Decitabine and Cisplatin are Synergistic to Exert Anti-Tumor Effect on Gastric Cancer via Inducing Sox2 DNA Demethylation. Onco Targets Ther 2021;14:623-36. [Crossref] [PubMed]
- Lund P, Kotova I, Kedinger V, et al. Transformation-dependent silencing of tumor-selective apoptosis-inducing TRAIL by DNA hypermethylation is antagonized by decitabine. Mol Cancer Ther 2011;10:1611-23. [Crossref] [PubMed]
- Fan X, Li Y, Yi X, et al. Epigenome-wide DNA methylation profiling of portal vein tumor thrombosis (PVTT) tissues in hepatocellular carcinoma patients. Neoplasia 2020;22:630-43. [Crossref] [PubMed]
- Zhu H, Huang M, Ren D, et al. The synergistic effects of low dose fluorouracil and TRAIL on TRAIL-resistant human gastric adenocarcinoma AGS cells. Biomed Res Int 2013;2013:293874. [Crossref] [PubMed]
- Zhou YY, Wang HY, Tang ZG, et al. Two new formulae for evaluating effectiveness of drug combinations and revision of Bürgi's and Jin's modified Bürgi's formula. Zhongguo Yao Li Xue Bao 1984;5:217-21.
- Suzuki M, Shigematsu H, Shivapurkar N, et al. Methylation of apoptosis related genes in the pathogenesis and prognosis of prostate cancer. Cancer Lett 2006;242:222-30. [Crossref] [PubMed]
- Li L, Fan B, Zhang LH, et al. Trichostatin A potentiates TRAIL-induced antitumor effects via inhibition of ERK/FOXM1 pathway in gastric cancer. Tumour Biol 2016;37:10269-78. [Crossref] [PubMed]
- Li L, Wen XZ, Bu ZD, et al. Paclitaxel enhances tumoricidal potential of TRAIL via inhibition of MAPK in resistant gastric cancer cells. Oncol Rep 2016;35:3009-17. [Crossref] [PubMed]
- Dyer MJ, MacFarlane M, Cohen GM. Barriers to effective TRAIL-targeted therapy of malignancy. J Clin Oncol 2007;25:4505-6. [Crossref] [PubMed]
- Rudner J, Jendrossek V, Lauber K, et al. Type I and type II reactions in TRAIL-induced apoptosis -- results from dose-response studies. Oncogene 2005;24:130-40. [Crossref] [PubMed]
- Karikari CA, Roy I, Tryggestad E, et al. Targeting the apoptotic machinery in pancreatic cancers using small-molecule antagonists of the X-linked inhibitor of apoptosis protein. Mol Cancer Ther 2007;6:957-66. [Crossref] [PubMed]
- Shetty S, Gladden JB, Henson ES, et al. Tumor necrosis factor-related apoptosis inducing ligand (TRAIL) up-regulates death receptor 5 (DR5) mediated by NFkappaB activation in epithelial derived cell lines. Apoptosis 2002;7:413-20. [Crossref] [PubMed]
- Zhou Q, Yuan J, Liu Y, et al. Cisatracurium besilate enhances the TRAIL-induced apoptosis of gastric cancer cells via p53 signaling. Bioengineered 2021;12:11213-24. [Crossref] [PubMed]
- Li H, Lv J, Guo J, et al. 5-Fluorouracil enhances the chemosensitivity of gastric cancer to TRAIL via inhibition of the MAPK pathway. Biochem Biophys Res Commun 2021;540:108-15. [Crossref] [PubMed]
- Zhao LM, Li L, Huang Y, et al. Antitumor Effect of Periplocin in TRAIL-Resistant gastric cancer cells via upregulation of death receptor through activating ERK1/2-EGR1 pathway. Mol Carcinog 2019;58:1033-45. [Crossref] [PubMed]
- Xie R, Gao CC, Yang XZ, et al. Combining TRAIL and liquiritin exerts synergistic effects against human gastric cancer cells and xenograft in nude mice through potentiating apoptosis and ROS generation. Biomed Pharmacother 2017;93:948-60. [Crossref] [PubMed]
- You C, Sun Y, Zhang S, et al. Trichosanthin enhances sensitivity of non-small cell lung cancer (NSCLC) TRAIL-resistance cells. Int J Biol Sci 2018;14:217-27. [Crossref] [PubMed]
- Han L, Dai S, Li Z, et al. Combination of the natural compound Periplocin and TRAIL induce esophageal squamous cell carcinoma apoptosis in vitro and in vivo: Implication in anticancer therapy. J Exp Clin Cancer Res 2019;38:501. [Crossref] [PubMed]
- Song P, Wu L, Guan W. Genome-Wide Identification and Characterization of DNA Methylation and Long Non-Coding RNA Expression in Gastric Cancer. Front Genet 2020;11:91. [Crossref] [PubMed]
- Kantarjian H, Oki Y, Garcia-Manero G, et al. Results of a randomized study of 3 schedules of low-dose decitabine in higher-risk myelodysplastic syndrome and chronic myelomonocytic leukemia. Blood 2007;109:52-7. [Crossref] [PubMed]
- Kantarjian H, Issa JP, Rosenfeld CS, et al. Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer 2006;106:1794-803. [Crossref] [PubMed]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev 2002;16:6-21. [Crossref] [PubMed]
- Xiao J, Liu P, Wang Y, et al. A Novel Cognition of Decitabine: Insights into Immunomodulation and Antiviral Effects. Molecules 2022;27:1973. [Crossref] [PubMed]
- Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A 2006;103:1412-7. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202-9. [Crossref] [PubMed]
- Lee KH, Lim SW, Kim HG, et al. Lack of death receptor 4 (DR4) expression through gene promoter methylation in gastric carcinoma. Langenbecks Arch Surg 2009;394:661-70. [Crossref] [PubMed]


