
Long non-coding RNA CNALPTC1 promotes gastric cancer progression by regulating the miR-6788-5p/PAK1 pathway
Highlight box
Key findings
• CNALPTC1 promotes GC development by negatively regulating the miR-6788-5p/PAK1 pathway. GC therapy may be improved by conducting targeted studies of the CNALPTC1/miR-6788-5p/PAK1 axis.
What is known and what is new?
• The aberrant expression of lncRNA is considered as an important regulatory factor of tumor development.
• CNALPTC1 expression in GC cell lines was identified and the related factors of its expression differences in combination with clinicopathological parameters were observed. The effects of upregulation and silencing of CNALPTC1 on proliferation, invasion, migration, and other biological functions of GC cell lines were observed.
What is the implication, and what should change now?
• It is necessary to explore novel biomarkers, investigate the fundamental mechanisms of onset and advancement of GC to improve the diagnosis and overall survival of GC patients.
Introduction
Gastric cancer (GC) is a prevalent gastrointestinal cancer worldwide and is caused by many factors. With the application of surgery, chemotherapy, immunotherapy, radiotherapy, and targeted molecular therapy, the survival and prognosis of GC patients have been greatly improved (1-3). However, the diagnosis of GC remains a major concern, with over 50% of patients being diagnosed after the tumor has spread, and the 5-year survival rate in most parts of the world is under 20% (4-6). Early-stage detection and treatment are vital for the reduction in mortality rates due to the presence of GC. Moreover, the detection of specific, sensitive, prognostic biomarkers and new therapeutically important targets is critical to the success of treatment. Currently, the underlying molecular mechanisms of the disease mostly remain unidentified; the discovery of novel biomarkers is a necessity to investigate the fundamental mechanisms of onset and advancement of GC in order to improve the diagnosis and overall survival of GC patients.
Eukaryotic cells encode long non-coding RNAs (lncRNAs) primarily transcribed via RNA polymerase II; the length of the transcripts is around 200 nucleotides and they normally lack a recognizable open reading frame. Previously published reports have revealed that lncRNAs are essential for gene expression regulation and chromatin structure at transcriptional, posttranscriptional, and epigenetic levels (7-9). Furthermore, the aberrant expression of lncRNAs is a leading cause of various diseases such as cancer; hence, they are considered important regulatory factors of tumor development (10-12). Previous research has confirmed that lncRNAs can contribute to the regulation of the epithelial-to-mesenchymal transition (EMT) process (13,14). The abnormal expression of lncRNAs can act as a possible biomarker for early detection, treatment, and prognosis of a variety of tumors (15,16). However, due to substantially large quantities and complex mechanisms, the functions of most lncRNAs in the onset, progression, and metastasis of GC have not been clarified.
LncRNA CNALPTC1 was first identified in papillary thyroid cancer (PTC), located on chromosome 1p366.22-p36.21 lncRNA ENST00000606790. It can stimulate the PTC cells’ proliferation and migration and inhibit cell apoptosis. Substantial upregulation of CNALPTC1 was observed in PTC in comparison to paracancerous tissues, and the increased expression was associated with invasive clinicopathological features. The study has been conducted in PTC to explore the biological role and possible mechanism of CNALPTC1, and the gene copy number amplification of the gene encoding ENST00000606790 (original serial number) was confirmed. Therefore, this lncRNA was named as copy number extended growth chain non-coding RNA 1 of PTC. The copy number amplified lncRNA CNALPTC1 promotes PTC progression via the sponging miR-30 family, which implies that CNALPTC1 may be a novel therapeutic target for PTC (17). Currently, CNALPTC1 has not been reported in other tumors, and its relationship and mechanism of action in the development of GC remain unclear, which requires further exploration. In our research, we identified CNALPTC1 expression in GC cell lines and observed the related factors of its expression differences in combination with clinicopathological parameters. The effects of upregulation and silencing of CNALPTC1 on proliferation, invasion, migration, and other biological functions of GC cell lines were observed. Bioinformatics was used to analyze and predict the microRNAs (miRNAs) targeted to CNALPTC1 and to examine CNALPTC1-related molecular mechanisms in the regulation and development of GC by predicting the target genes interacting with miRNA. We present the following article in accordance with the MDAR reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1069/rc).
Methods
Clinicopathological samples
In this study, 80 patients with GC underwent radical surgery in the Third Department of General Surgery of the Fourth Hospital of Hebei Medical University between May 2018 and July 2019. After obtaining the tumor tissues and adjacent mucosa tissues (more than 5 cm from the tumor edge), each tissue was divided into two parts. Some the fresh tissue was immediately stored in liquid nitrogen and stored in a −80 ℃ refrigerator for later experimental detection. The other parts were confirmed by routine pathological examination by the experienced pathologist as GC, and paracancerous tissue histopathology was carried out for the purpose of confirming that there were no tumor cells and mucosal glandular dysplasia in normal tissues. The patients included in this study had not received any of the following treatments: preoperative radiotherapy, chemotherapy, targeted or biological immunotherapy, and had no history of other malignant tumors. The Ethics Committee of the Fourth Hospital of Hebei Medical University (No. 2020ky250) granted its approval for this research, and informed consent was provided by all participants. The study was conducted with the Declaration of Helsinki (as revised in 2013).
Cell cultures and transfection
The Cell Bank of the Chinese Academy of Sciences (Shanghai, China) was contacted to purchase various cell lines, including healthy human gastric mucous epithelium cell line (GES-1) and GC cell lines, namely, MKN74, MKN45, and AGS. The culturing of all the cells was conducted at a temperature of 37 ℃ with 5% CO2 in Roswell Park Memorial Institute (RPMI)-1640 medium (Gibco, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, MA, USA), penicillin 100 U/mL, and streptomycin (Invitrogen, Carlsbad, CA, USA) 100 mg/mL.
Lipofectamine 2000 (Invitrogen) was employed to conduct the transfection of small interfering RNAs (siRNAs) and plasmid vectors into GC cells, according to the transfection reagent operation instructions. The siRNA (1 of the 3 individual CNALPTC1 siRNAs was selected after validation) was used for CNALPTC1 knockdown in MKN45 cells, whereas the plasmid pcDNA3.1-CNALPTC1 was utilized for CNALPTC1 overexpression in MKN74 cells. The miR-6788-5p mimic, miR-6788-5p inhibitor, and negative control (NC or vector) were purchased from Invitrogen. The downregulation of CNALPTC1 was achieved by using Lipofectamine 2000 to conduct transfection of MKN74 cells with the NC siRNA’s target sequence. The number of coding sequences was increased as per the manufacturer’s protocol to achieve overexpression of CNALPTC1, and subcloning was performed in the pcDNA3.1 (+) vector (Invitrogen). Furthermore, the transfection of MKN74 cells with a Blank control (NC vector) or a CNALPTC1-expressed plasmid was carried out with the aid of Lipofectamine 2000. The transfection process was completed over 48 hours, and the cells were isolated to perform quantitative real-time polymerase chain reaction (qRT-PCR) and western blot assays. The siRNA sequences as follows: siRNA#1 5'-GGAGAAUGCAGAAUGGUCAGACUAC-3'; siRNA#2 5'-GCAGGAACUUGAUUACUUUGAGUGC-3'; siRNA#3 5'- ACACCAUUAGGCAUAGAUUCAGUGT-3'.
RNA extraction and qRT-PCR
The total RNA was extracted from tissues and cell lines utilizing RNAiso Plus (TaKaRa, Tokyo, Japan) following the protocol provided by the manufacturer. The purity of RNA was evaluated by optical density (OD)260/OD280 ratio. A Transcriptor cDNA Synth Kit (Roche, Basel, Switzerland) was employed to conduct reverse transcription as per the instructions provided by the manufacturer. A TransStart Top Green qPCR SuperMix (TransGen, Beijing, China) was utilized to conduct qRT-PCR by employing a RT-PCR system (ABI 7500; Shanghai Simenbio Technology Ltd., Shanghai, China). The relative level of expression of all the target genes was evaluated by employing the 2−ΔΔCt relative quantification method. Furthermore, glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was employed as an internal control of lncRNA and messenger RNA (mRNA), whereas U6 was employed as an internal control for miRNA. The primer sequences utilized in the experiment are as follows in Table 1.
Table 1
| Gene | Primer sequence |
|---|---|
| CNALPTC1 | F: 5'-CACTTTGAACTGGGGCTG-3' |
| R: 5'-GGAACTGTAATCTCACTGGACAA-3' | |
| GAPDH | F: 5'-AATCCCATCACCATCTTCCAG-3' |
| R: 5'-CCTTCTCCATGGTGGTGAAGAC-3' | |
| PCNA | F: 5'-GTCTCAATTGCG-TTACTATGTC-3' |
| R: 5'-TAATAGCATCATAATCTTGATCTG-3' | |
| p16 | F: 5'-GAAGAAAGAGGAGGGGTTGG-3' |
| R: 5'-CAGCACTCTTAG-G-AACCTCTCAT-3' | |
| p21 | F: 5'-TGCAACTACTACAGAAACTGCTG-3' |
| R: 5'-CAAAGTGGTCGGTAGCCACA-3' | |
| CyclinD1 | F: 5'-CAAGGCCTGAACCTGAGGAG-3' |
| R: 5'-CTTGGGGTCCATGTTCTGCT-3' | |
| CyclinE | F: 5'-ATGCCGAGGGAGCGCAGGGAG-3' |
| R: 5'-GGTCTCCTATGAAGTTTATAGAC-3' | |
| MMP2 | F: 5'-CCAACTACAACTTCTTCCCTCG-3' |
| R: 5'-TCACATCGCTCCAGACTTG-3' | |
| MMP9 | F: 5'-ACGCAGACATCGTCATCCA-3' |
| R: 5'-AGGGACCACAACTCGTCATC-3' | |
| TIMP1 | F: 5'-TTCTGGCATCCTGTTGTTG-3' |
| R: 5'-GTGGTCTGGTTGACTTCTGG-3' | |
| TIMP2 | F: 5'-CGACATTTATGGCAACCCT-3' |
| R: 5'-ATTCCTTCTTTCCTCCAACG-3' | |
| NM23 | F: 5'-AAGGAGATCGGCTTGTGGTTT-3' |
| R: 5'-CTGAGCACAGCTCGTGTAATC-3' | |
| ICAM1 | F: 5'-GTCATCATCACTGTGGTAGCAG-3' |
| R: 5'-GGCTTGTGTGTTCGGTTTC-3' | |
| E-cadherin | F: 5'-GTGGTCAAAGAGCCCTTACTG-3' |
| R: 5'-CGTTACGAGTCACTTCAGGC-3' | |
| N-cadherin | F: 5'-TCATTGCCATCCTGCTCTG-3' |
| R: 5'-CATCCATACCACAAACATCAGC-3' | |
| Vimentin | F: 5'-AAATGGCTCGTCACCTTCG-3' |
| R: 5'-AGAAATCCTGCTCTCCTCGC-3' | |
| Snail | F: 5'-TCGGAAGCCTAACTACAGCG-3' |
| R: 5'-CAGAGTCCCAGATGAGCATTG-3' | |
| Twist | F: 5'-GTCCGCAGTCTTACGAGGAG-3' |
| R: 5'-GCTTGAGGGTCTGAATCTTGCT-3' | |
| ZEB1 | F: 5'-GCTTCTCACACTCTGGGTCTTA-3' |
| R: 5'-CCTCATTCTCTGCCTCTTCTACC-3' | |
| β-actin | F: 5'-TACCTCATGAAGATCCTCACCGA-3' |
| R: 5'-AAGCATTTGCGGTGGACGAT-3' | |
| MiR-6788-5p | F: 5'-CTGGGAGAA-GAGTGGTGAAGA-3' |
| R: 5'-CUGGGAGAAGAGUGGUGAAGA-3' | |
| PAK1 | F: 5'-AGCTAACTGACTTTGGATTC-3' |
| R: 5'-GGGTTTTCATTGAG-GTATGG-3' |
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
The viability of transfected MKN45 and MKN74 cells with si-CNALPTC1 and pcDNA3.1-CNALPTC1, respectively, was calculated with the aid of the MTT Cell Proliferation and Cytotoxicity Assay Kit (Solarbio, Beijing, China). Trypsin was utilized to detach cells in the logarithmic growth phase, and the cell suspension was prepared after performing transfection. Subsequently, the cells were seeded at 3×105 cells/well onto 96-well plates, with 200 µL medium added in all the wells and marked, and incubation was performed for 24 hours with 5% CO2 at a temperature of 37 ℃. After incubation, the removal of suspension was performed, and the solution was replaced with 200 µL dimethyl sulfoxide (DMSO; BioSharp, Hefei, China) for substrate dissolution for 10 minutes. The absorbance was evaluated using a microplate spectrophotometer (Bio-Rad, Hercules, CA, USA) at 490 nm.
Flow cytometry
Lipofectamine 2000 was employed to conduct the transfection of MKN45 and MKN74 cells with si-CNALPTC1 and pcDNA3.1-CNALPTC1, respectively, and the transfected cells were obtained after 24 hours. Moreover, the harvesting and resuspension of transfected cells were conducted in the binding buffer (1×), followed by the incubation with propidium iodide (PI) by employing the DNA content quantitative measurement method (cell cycle) (Solarbio), which was identified by using FACScan (Becton Dickinson, San Jose, CA, USA). The count and comparison of cell percentage in the G0/G1, S, and G2/M phases were performed.
Wound-healing assay
The transfected cells were placed in a 6-well plate at a density of 5×105 cells. A wound line was scratched vertically to the bottom by utilizing a pipette tip of 10 µL until the cells were grown to 100% confluence. Moreover, phosphate-buffered saline (PBS) was used to rinse off the floating cells, and the adherent cells were cultured for 24 hours using Dulbecco’s modified Eagle medium (DMEM) medium containing 1% FBS. An inverted optical microscope (Olympus, Tokyo, Japan) was employed for the observation of the wound closure at 0 and 24 hours.
Transwell assay
After transfection, the treated cells were placed in a 24-well plate at the density of 5×105 cells. Subsequently, seeding of cells was performed in the upper chamber (Sigma-Aldrich, St. Louis, MO, USA) by using 50 µL Matrigel-coated membranes, followed by adding 600 µL culture medium with 10% FBS, as a chemoattractant, for a period of 24 hours at 37 ℃ containing 5% CO2 to the lower chamber. The non-invaded cells were removed and then stained using 0.1% of crystal violet. The cell imaging and counting were performed using an inverted optical microscope.
Western blot
The western blot method was employed for the analysis of the protein expression. After performing various treatments, cell lysis was carried out using radioimmunoprecipitation assay (RIPA) buffer (Solarbio), which comprises a combination of phosphatase and protease inhibitors (Roche) and quantified by employing the BAC kit (Solarbio). Approximately 30 µg of proteins were loaded in this experiment, and separation was performed using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The transfer of proteins from SDS-PAGE to polyvinylidene fluoride (PVDF) membrane was performed, and its blocking was carried out using 5% of nonfat milk in tris-buffered saline with Tween 20 (TBST) solution. The protein recognition procedure was performed by utilizing corresponding primary antibodies incubated overnight at a temperature of 4 ℃ and secondary antibodies at room temperature for a duration of 2 hours. Furthermore, incubation of the membrane with proteins was carried out in the enhanced chemiluminescence (ECL) solution, and the Bio-Rad chemiluminescence system was utilized to display the signals. All the antibodies (1:2,000 dilutions) were provided by Abcam (Cambridge, UK).
Luciferase reporter assay
The PCR amplification was carried out of the 3'-untranslated region (3'-UTR) of lncRNA CNALPTC1 comprising the predicted miR-6788-5p binding site, followed by cloning into a Dual-Luciferase miRNA Target Expression Vector (Promega, Madison, WI, USA) to produce CNALPTC1-wild-type (CNALPTC1-Wt). A similar method was adopted for the production of CNALPTC1-mutated-type (CNALPTC1-Mut), PAK1-wild-type (PAK1-Wt), or PAK1-mutated-type (PAK1-Mut) constructs. As per the protocol, the calculation of luciferase activity was carried out with the aid of the dual-luciferase reporter assay system (Promega) after incubating for 48 hours. MKN45 cells were transfected. Three separate studies were carried out, and the corresponding activity of luciferase was normalized to the Renilla luciferase internal control.
Statistical analysis
The software GraphPad Prism 8.0 (GraphPad Software, San Diego, CA, USA) was for statistical analyses, and the mean ± standard deviation was employed for the expression of data. One-way analysis of variance (ANOVA) was utilized for statistical comparison, and P<0.05 was taken as a statistically significant difference.
Results
CNALPTC1 is up-regulated in human GC tissues and cell lines
The expression of CNALPTC1 was different in various stages of GC, namely, the expression level was higher in the advanced stages of GC in contrast with that in the early stages of GC, according to The Cancer Genome Atlas (TCGA) datasets and Genotype-Tissue Expression (GTEx) database (Figure 1A). Moreover, the function of CNALPTC1 in human GC tissues was investigated by studying the CNALPTC1 expression in GC and neighboring healthy tissues.

When compared to the adjoining healthy tissues, we observed that CNALPTC1 was expressed at a remarkably higher level in GC tissues (Figure 1B) as well as the GC cell lines (MKN74, AGS, and MKN45) in comparison to the healthy GC line GES-1. Among various differentiated GC cell lines, the expression level was highest in the poorly differentiated MKN45 cell line and lowest in the well-differentiated MKN74 cell line, indicating that the worse the differentiation, the higher the expression level of lncRNA CNALPTC1 (Figure 1C). Furthermore, to analyze the clinical value of CNALPTC1 overexpression in GC, we investigated its expression in 80 patients with GC and assessed the correlation with clinicopathological characteristics. The median expression level of CNALPTC1 was considered a key value. The GC tissue was divided into two groups: low-expression and high-expression groups, with 37 and 43 cases in each group, respectively. The expression levels of CNALPTC1 in cancer tissue revealed a positive correlation with Lauren classification (P=0.001), tissue differentiation (P=0.001), tumor-node-metastasis (TNM) stage (P=0.035), infiltrating depth (P=0.019), nodal status (P=0.003), and blood vessel invasion (P=0.004), but it did not display any link to other clinicopathological parameters such as age, gender, cancer size and location, distant metastases, and human epidermal growth factor receptor 2 (HER2) amplified state (Table 2).
Table 2
| Clinicopathological parameters | N | Expression of lncRNA CNALPTC1, n | χ2 | P value | |
|---|---|---|---|---|---|
| Higher expression | Lower expression | ||||
| Gender | 0.302 | 0.583 | |||
| Male | 48 | 27 | 21 | ||
| Female | 32 | 16 | 16 | ||
| Age (years) | 0.409 | 0.522 | |||
| ≤57 | 38 | 19 | 19 | ||
| >57 | 42 | 24 | 18 | ||
| Tumor location | 1.863 | 0.686 | |||
| Upper third | 53 | 30 | 23 | ||
| Middle third | 14 | 7 | 7 | ||
| Lower third | 12 | 6 | 6 | ||
| Full stomach | 1 | 0 | 1 | ||
| Tumor size (cm) | 1.923 | 0.165 | |||
| ≤5.1 | 52 | 25 | 27 | ||
| >5.1 | 28 | 18 | 10 | ||
| Lauren classification | 14.856 | 0.001* | |||
| Intestinal | 45 | 16 | 29 | ||
| Diffuse | 20 | 17 | 3 | ||
| Mixed | 15 | 10 | 5 | ||
| Tissue differentiation | 12.645 | 0.001* | |||
| Well | 5 | 0 | 5 | ||
| Moderate | 41 | 18 | 23 | ||
| Poor | 34 | 25 | 9 | ||
| Infiltrating depth | 9.088 | 0.019* | |||
| T1 | 3 | 0 | 3 | ||
| T2 | 27 | 10 | 17 | ||
| T3 | 43 | 28 | 15 | ||
| T4 | 7 | 5 | 2 | ||
| Nodal status | 13.918 | 0.003* | |||
| N0 | 20 | 5 | 15 | ||
| N1 | 17 | 7 | 10 | ||
| N2 | 18 | 12 | 6 | ||
| N3 | 25 | 19 | 6 | ||
| Distant metastases | 1.765 | 0.497 | |||
| M0 | 78 | 41 | 37 | ||
| M1 | 2 | 2 | 0 | ||
| TNM stage | 7.723 | 0.035* | |||
| I | 6 | 2 | 4 | ||
| II | 30 | 12 | 18 | ||
| III | 42 | 28 | 14 | ||
| IV | 2 | 1 | 1 | ||
| Blood vessel invasion | 8.177 | 0.004* | |||
| Yes | 18 | 15 | 3 | ||
| No | 62 | 28 | 34 | ||
| HER2 | 0.013 | 1.000 | |||
| Positive | 9 | 5 | 4 | ||
| Negative | 71 | 38 | 33 | ||
*P<0.05. LncRNA, long non-coding RNA; GC, gastric cancer; TNM, tumor-node-metastasis; HER2, human epidermal growth factor receptor 2.
Downregulation of CNALPTC1 suppresses GC cell proliferation, migration, and invasion
The level of expression of CNALPTC1 in the undifferentiated MKN45 GC cell line was the highest among various GC cell lines. Therefore, this cell line was selected for the inhibition experiment. The siRNA with the highest knockout rate was selected for subsequent experiments, which revealed that CNAlPTC1-siRNA can effectively inhibit the expression of endogenous CNALPTC1 in MKN45 GC cells (Figure 2A). Cell proliferation assay (MTT) findings revealed that CNALPTC1 knockdown has considerable inhibitory effects on the growth of cells (Figure 2B). After CNALPTC1 expression was inhibited, the expression level of proliferative factor proliferating cell nuclear antigen (PCNA) in MKN45 GC cells was significantly decreased (Figure 2C). Flow cytometry findings revealed that CNALPTC1 knockdown elevated the cell percentage in G0/G1 phase, and that a reduction in the cell percentage in S and G2/M phases was observed. Moreover, the level of expression of cell cycle-related genes p16 and p21 considerably improved, and the levels of expression of cyclinD1 and cyclinE were significantly decreased (Figure 2D). Additionally, the transwell and wound-healing experiments revealed that CNALPTC1 knockdown substantially suppressed cell invasion and migration in MKN45 GC cells (Figure 2E). The levels of expression of ICAM1, NM23, MMP2, MMP9, cell invasion- and migration-related genes were lowered, whereas no significant changes were observed in TIMP1 and TIMP2 (Figure 2F).
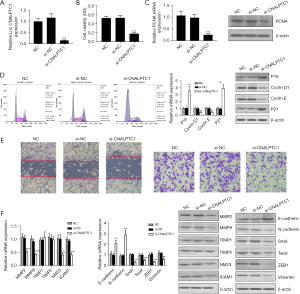
Upregulation of CNALPTC1 enhances GC cell proliferation, migration, and invasion
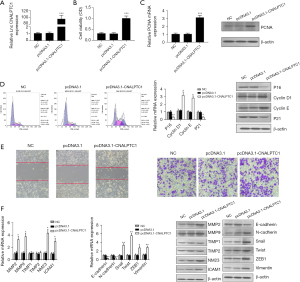
The expression level of CNALPTC1 in the well-differentiated MKN74 GC cell line was the lowest among the GC cell lines with different degrees of differentiation. Therefore, the highly differentiated MKN74 GC cell line was selected for the overexpression experiment to construct the PCDNA3.1-CNALPTC1 overexpression plasmid. After transfection of PCDNA3.1-CNALPTC1 and PCDNA3.1-empty plasmids into MKN74 cells, the expression level of CNALPTC1 in MKN74 cells of the PCDNA3.1-CNALPTC1 group was significantly increased indicating that overexpression treatment had been achieved (Figure 3A). The cell proliferation assay (MTT) findings revealed that overexpression of CNALPTC1 remarkably promoted cell proliferation (Figure 3B). The overexpression of CNALPTC1 resulted in the overexpression of proliferation factor PCNA in MKN74 GC cells (Figure 3C). The findings of flow cytometry revealed that the cell count in G0/G1 phase reduced considerably after overexpression of CNALPTC1, whereas the cell number in S and G2/M phases increased substantially. The expression levels of p16 and p21, the cell cycle-related genes, were suppressed substantially, whereas an upregulation in the levels of expression of cyclinD1 and cyclinE was observed (Figure 3D). Additionally, transwell and wound healing analyses revealed that CNALPTC1 upregulation significantly suppressed the invasion and migration of MKN74 GC cells (Figure 3E). The levels of expression of invasion- and migration related-genes such as NM23, MMP2, ICAM1, and MMP9 increased, whereas TIMP1 and TIMP2 expression levels did not change significantly (Figure 3F).
CNALPTC1 induces EMT in GC cells
As tumor metastasis is influenced by the key factor EMT, the evaluation of the effects of CNALPTC1 on EMT was carried out by employing qRT-PCR and western blot. The assessment of the expression of EMT-related markers after upregulation or downregulation of CNALPTC1 was performed. E-cadherin and N-cadherin overexpression was observed after CNALPTC1 expression was downregulated, whereas the expressions of Snail, Twist, ZEB1, and Vimentin were significantly reduced, suggesting that CNALPTC1 knockdown led to inhibition of the migration and invasion of GC MKN45 cells (Figure 2F). Contrary findings were observed in GC MKN74 cells due to overexpression of CNALPTC1 (Figure 3F). These findings indicated that overexpression of CNALPTC1 can induce migration and invasion in GC cells.
CNALPTC1 stimulates GC progression by regulating the miR-6788-5p/PAK1 pathway
The identification of the function of CNALPTC1 in GC was conducted by a comprehensive investigation of its target miRNAs and utilizing the online software “miRDB” (https://mirdb.org/). The obtained data showed that CNALPTC1 had a putative target sequence for miR-6788-5p. Furthermore, the results indicated that miR-6788-5p and lncRNA CNALPTC1 had a maximum of 8 consecutive bases for a complementary pairing. In order to confirm these predictions, luciferase reporter analysis was performed with CNALPTC1 wild-type reporter vector (CNALPTC1-Wt) containing the target sequence for miR-6788-5p and the mutation of seeded sequence (CNALPTC1-Mut) (Figure 4A). The findings indicated that overexpression of CNALPTC1 played a significant role in inhibiting miR-6788-5p expression in MKN74 cells (Figure 4B). Moreover, the inhibition of CNALPTC1 expression in MKN45 cells resulted in overexpression of miR-6788-5p, suggesting that the expression of CNALPTC1 displayed a negative correlation with miR-6788-5p (Figure 4C). Subsequent luciferase assays revealed that the co-transfection of MKN45 cells with CNALPTC1-Wt and miR-6788-5p mimic suggested lower luciferase activity in comparison to the cells co-transfected with miR-NC mimic, whereas the mutant luciferase activity of CNALPTC1 did not alter significantly. The results suggested that the targeted sequence mutation clearly eliminated the impact of miR-6788-5p upregulation on reporter gene expression (Figure 4D).

The miRNA target prediction databases such as miRDB and TargetScan (https://www.targetscan.org/vert_80/) were utilized to predict miR-6788-5p targets. The data indicated the existence of a putative binding site for miR-6788-5p in the 3'-UTR of PAK1, which was verified by conducting the luciferase reporter analysis of PAK1 3'-UTR reporter vector (PAK1-Wt) containing the putative miR-6788-5p-binding site or the mutation of seeded sequence (PAK1-Mut) (Figure 4E). The findings revealed that the transient introduction of miR-6788-5p mimic significantly suppressed the expression of PAK1 in MKN45 cells, in comparison to the NC (Figure 4F). Additionally, the inhibition of miR-6788-5p in MKN74 cells (transfected miR-6788-5p inhibitor), resulted in the overexpression of PAK1 (Figure 4G). The co-transfection of miR-6788-5p mimic and PAK1-Wt into MKN45 cells induced lower luciferase activity in comparison to the cells co-transfected with miR-NC mimic, whereas the mutant luciferase activity of PAK1 did not change significantly (Figure 4H). All these data indicated that PAK1 was a functional target of miR-6788-5p in GC cells.
The expression of genes is regulated by lncRNAs serving as miRNAs’ molecular sponge. Furthermore, we examined the function of CNALPTC1 as a modulator of PAK1 expression in GC cells. Overexpression of CNALPTC1 in GC MKN74 cells up-regulated the expression level of PAK1. After co-transfection with pcDNA3.1-CNALPTC1 and miR-6788-5p mimic, the level of PAK1 mRNA and protein was restored (Figure 4I). Silencing of CNALPTC1 silencing substantially decreased the levels of PAK1 expression as compared to the NC. However, co-transfection with si-CNALPTC1 and miR-6788-5p inhibitor restored the level of PAK1 mRNA and protein expression (Figure 4J). This data revealed the function of CNALPTC1 as miR-6788-5p’s molecular sponge to regulate PAK1 expression, thereby playing an essential part in the advancement of GC.
Discussion
Although lncRNAs have no protein-coding function, they can participate in regulatory processes, such as chromatin modification, transcription interference, selective shearing, transcription activation, intracellular transport, and regulation of proto-oncogene activation, thus regulating gene expression at multiple levels (18). Some lncRNAs have similar expression changes and functions in different malignant tumors, and some others have different expressions and functions in different tumors (19). The possible function of lncRNAs in regulating malignant manifestations of tumor cells has attracted widespread attention (20), and they play a significant regulatory function in tumor proliferation, migration, and invasion. Abnormal expression of lncRNAs has been observed in different tumors, including prostate cancer, esophageal cancer, cervical cancer, cholangiocarcinoma, and so on (21,22). Furthermore, lncRNAs are related to the transduction process of multiple signaling pathways (23) and act as a “sponge” or bait and indirectly affect the expression of miRNA target genes by negatively regulating their expression with miRNA (24,25).
Treatments for advanced GC are limited but have the potential to improve survival and quality of life. It is important to further study the pathogenesis, new treatments, and prognostic markers of GC. Numerous lncRNAs have been identified that play a vital role in GC development. In this study, we found the GC-related lncRNA CNALPTC1, and the findings revealed CNALPTC1 overexpression in GC tissues and cells.
The elevated expression level of CNALPTC1 was associated with Lauren classification, depth of invasion, TNM stage, lymph node metastasis, degree of tissue differentiation, and vascular invasion, suggesting that abnormal expression of CNALPTC1 may be linked to the GC progression. A published study on CNALPTC1 has shown that it can induce the proliferation and migration of PTC cells and result in the suppression of cell apoptosis (17). In this study, the effects of knockdown and upregulation of CNALPTC1 on the biological functions of GC cells were analyzed. The results revealed that CNALPTC1 knockdown reduced the migration, growth, invasion, and EMT process of GC cells, whereas upregulation of CNALPTC1 can promote the migration, growth, invasion, and EMT process of GC cells.
LncRNAs have important biological functions as tumor-competing endogenous RNAs (ceRNAs). Moreover, bioinformatics tools were used to analyze and construct lncRNA-miRNA-mRNA ceRNA networks and identify potential lncRNA biomarkers in GC (25). The regulatory mechanism and function of lncRNA in GC have not been fully elucidated. Currently, only a few reports have been published about CNALPTC1, and its mechanism in the development of GC is still not clear. In this study, bioinformatics was utilized to analyze and predict miR-6788-5p targeted to CNALPTC1 and to study the molecular mechanism of the progression and occurrence of GC by predicting the target gene PAK1 interacting with miRNA. It was found that upregulation of CNALPTC1 inhibited miR-6788-5p expression in GC cells, and suppression of CNALPTC1 elevated miR-6788-5p expression. Luciferase assay results confirmed that miR-6788-5p could target CNALPTC1, which could negatively regulate the expression of miR-6788-5p. PAK1 is the downstream target protein of Cdc42 and Rac1 of the Rho family of small G proteins and is also an essential regulator of cell migration and invasion (26-28). PAK1 plays a significant role in regulating cell death and survival signaling pathways by controlling apoptosis (29-31). Low expression of PAK1 leads to GC cell cycle arrest, and CDK4/6 inhibitors reduce GC cell viability and PAK1 expression (32); moreover, it is a target gene predicted by bioinformatics. Our research revealed that PAK1 expression in GC cells was decreased by up-regulating miR-6788-5p, and the expression of PAK1 was increased by inhibiting miR-6788-5p. Luciferase assay results confirmed that PAK1 is a target for miR-6788-5p, which can negatively regulate PAK1 expression.
It was confirmed that the invasion and growth capacities of GC cells are induced by CNALPTC1 by regulation of the expression of the miR-6788-5p/PAK1 axis. Our work has laid a foundation for future research on the molecular mechanism of CNALPTC1 in the progression of GC.
Conclusions
We have identified that lncRNA CNALPTC1 was subjected to upregulation in GC tissues as well as in the cell lines observed in the in vitro study. The expression of PAK1 was positively correlated with CNALPTC1, which could target miR-6788-5p and play a role in its expression regulation, and it was found that miR-6788-5p regulated the expression of PAK1 in GC cells. In conclusion, our results indicated that the CNALPTC1/miR-6788-5p/PAK1 axis plays a significant role in GC, which provides potential molecular biomarkers and novel therapeutic targets for GC.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1069/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1069/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1069/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The Ethics Committee of the Fourth Hospital of Hebei Medical University (No. 2020ky250) granted its approval for this research, and informed consent was provided by all participants. The study was conducted with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Li W, Ng JM, Wong CC, et al. Molecular alterations of cancer cell and tumour microenvironment in metastatic gastric cancer. Oncogene 2018;37:4903-20. [Crossref] [PubMed]
- Liu HT, Ma RR, Lv BB, et al. LncRNA-HNF1A-AS1 functions as a competing endogenous RNA to activate PI3K/AKT signalling pathway by sponging miR-30b-3p in gastric cancer. Br J Cancer 2020;122:1825-36. [Crossref] [PubMed]
- Smyth EC. Chemotherapy for resectable microsatellite instability-high gastric cancer? Lancet Oncol 2020;21:204. [Crossref] [PubMed]
- Zheng G, Sundquist K, Sundquist J, et al. Second Primary Cancers After Gastric Cancer, and Gastric Cancer as Second Primary Cancer. Clin Epidemiol 2021;13:515-25. [Crossref] [PubMed]
- Högner A, Moehler M. Immunotherapy in Gastric Cancer. Curr Oncol 2022;29:1559-74. [Crossref] [PubMed]
- Katsushima K, Lee B, Kunhiraman H, et al. The long noncoding RNA lnc-HLX-2-7 is oncogenic in Group 3 medulloblastomas. Neuro Oncol 2021;23:572-85. [Crossref] [PubMed]
- Pan T, Yu Z, Jin Z, et al. Tumor suppressor lnc-CTSLP4 inhibits EMT and metastasis of gastric cancer by attenuating HNRNPAB-dependent Snail transcription. Mol Ther Nucleic Acids 2021;23:1288-303. [Crossref] [PubMed]
- Luo M, Liang C. LncRNA LINC00483 promotes gastric cancer development through regulating MAPK1 expression by sponging miR-490-3p. Biol Res 2020;53:14. [Crossref] [PubMed]
- Xiong HG, Li H, Xiao Y, et al. Long noncoding RNA MYOSLID promotes invasion and metastasis by modulating the partial epithelial-mesenchymal transition program in head and neck squamous cell carcinoma. J Exp Clin Cancer Res 2019;38:278. [Crossref] [PubMed]
- Li Q, Sun H, Luo D, et al. Lnc-RP11-536 K7.3/SOX2/HIF-1α signaling axis regulates oxaliplatin resistance in patient-derived colorectal cancer organoids. J Exp Clin Cancer Res 2021;40:348. [Crossref] [PubMed]
- Zhao J, Wu J, Qin Y, et al. LncRNA PVT1 induces aggressive vasculogenic mimicry formation through activating the STAT3/Slug axis and epithelial-to-mesenchymal transition in gastric cancer. Cell Oncol (Dordr) 2020;43:863-76. [Crossref] [PubMed]
- Sakai S, Ohhata T, Kitagawa K, et al. Long Noncoding RNA ELIT-1 Acts as a Smad3 Cofactor to Facilitate TGFβ/Smad Signaling and Promote Epithelial-Mesenchymal Transition. Cancer Res 2019;79:2821-38. [Crossref] [PubMed]
- Liu ZQ, He WF, Wu YJ, et al. LncRNA SNHG1 promotes EMT process in gastric cancer cells through regulation of the miR-15b/DCLK1/Notch1 axis. BMC Gastroenterol 2020;20:156. [Crossref] [PubMed]
- Zhang L, Niu H, Yang P, et al. Serum lnc34a is a potential prediction biomarker for bone metastasis in hepatocellular carcinoma patients. BMC Cancer 2021;21:161. [Crossref] [PubMed]
- Jin Z, Chen B. LncRNA ZEB1-AS1 Regulates Colorectal Cancer Cells by MiR-205/YAP1 Axis. Open Med (Wars) 2020;15:175-84. [Crossref] [PubMed]
- Chen C, Zhou L, Wang H, et al. Long noncoding RNA CNALPTC1 promotes cell proliferation and migration of papillary thyroid cancer via sponging miR-30 family. Am J Cancer Res 2018;8:192-206. [PubMed]
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 2009;23:1494-504. [Crossref] [PubMed]
- Chandra Gupta S, Nandan Tripathi Y. Potential of long non-coding RNAs in cancer patients: From biomarkers to therapeutic targets. Int J Cancer 2017;140:1955-67. [Crossref] [PubMed]
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 2014;15:7-21. [Crossref] [PubMed]
- Bill M, Papaioannou D, Karunasiri M, et al. Expression and functional relevance of long non-coding RNAs in acute myeloid leukemia stem cells. Leukemia 2019;33:2169-82. [Crossref] [PubMed]
- Hosono Y, Niknafs YS, Prensner JR, et al. Oncogenic Role of THOR, a Conserved Cancer/Testis Long Non-coding RNA. Cell 2017;171:1559-72.e20. [Crossref] [PubMed]
- Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017;36:5661-7. [Crossref] [PubMed]
- Paci P, Colombo T, Farina L. Computational analysis identifies a sponge interaction network between long non-coding RNAs and messenger RNAs in human breast cancer. BMC Syst Biol 2014;8:83. [Crossref] [PubMed]
- Kartha RV, Subramanian S. Competing endogenous RNAs (ceRNAs): new entrants to the intricacies of gene regulation. Front Genet 2014;5:8. [Crossref] [PubMed]
- Wang S, Wang SY, Du F, et al. Knockdown of PAK1 Inhibits the Proliferation and Invasion of Non-Small Cell Lung Cancer Cells Through the ERK Pathway. Appl Immunohistochem Mol Morphol 2020;28:602-10. [Crossref] [PubMed]
- Yang Z, Wang H, Xia L, et al. Overexpression of PAK1 Correlates with Aberrant Expression of EMT Markers and Poor Prognosis in Non-Small Cell Lung Cancer. J Cancer 2017;8:1484-91. [Crossref] [PubMed]
- Balasenthil S, Sahin AA, Barnes CJ, et al. p21-activated kinase-1 signaling mediates cyclin D1 expression in mammary epithelial and cancer cells. J Biol Chem 2004;279:1422-8. [Crossref] [PubMed]
- Shrestha Y, Schafer EJ, Boehm JS, et al. PAK1 is a breast cancer oncogene that coordinately activates MAPK and MET signaling. Oncogene 2012;31:3397-408. [Crossref] [PubMed]
- Wang Q, Li Z, Hu Y, et al. Circ-TFCP2L1 Promotes the Proliferation and Migration of Triple Negative Breast Cancer through Sponging miR-7 by Inhibiting PAK1. J Mammary Gland Biol Neoplasia 2019;24:323-31. [Crossref] [PubMed]
- Chung JH, Kim T, Kang YJ, et al. PAK1 as a Potential Therapeutic Target in Male Smokers with EGFR-Mutant Non-Small Cell Lung Cancer. Molecules 2020;25:5588. [Crossref] [PubMed]
- Fu H, Zhang W, Yuan Q, et al. PAK1 Promotes the Proliferation and Inhibits Apoptosis of Human Spermatogonial Stem Cells via PDK1/KDR/ZNF367 and ERK1/2 and AKT Pathways. Mol Ther Nucleic Acids 2018;12:769-86. [Crossref] [PubMed]
(English Language Editor: J. Jones)


