
Comprehensive analysis of an autophagy-related prognostic model for predicting survival based on TCGA and ICGC database in hepatocellular carcinoma patients
Highlight box
Key findings
• An autophagy-related gene (ARG) prognostic model of two key genes for predicting survival in hepatocellular carcinoma (HCC) patients.
What is known and what is new?
• Studies on multi-gene prognostic markers for HCC have been reported.
• The two ARGs included in the prognostic models had not been reported previously, and the differentially expressed ARGs may provide a new perspective for the study of HCC molecular mechanisms.
What is the implication, and what should change now?
• Relying solely on ARGs for prediction is not very accurate, and ARGs need to be combined with other factors to ensure comprehensive prediction.
Introduction
Hepatocellular carcinoma (HCC) is one of the most prevalent cancers in the world, and carries heavy clinical, financial, and psychological costs (1). Early-stage HCC patients may benefit from liver resection, ablation, and transplantation; however, the majority of HCC patients are diagnosed at intermediate and advanced stages and thus have only a few treatment options (2,3). Systemic therapy, coupled with targeted sellers and immune checkpoint inhibitors, continues to be essential for advanced stage HCC (4). However, patients with HCC frequently have a very poor prognosis due to recurrence and chemoresistance. Following advances in multi-omics profiling, research has produced prognostic candidates for clinical software (5,6). To predict the prognosis of HCC patients, strong molecular indicators need to be identified.
The prognosis of HCC patients is very unpredictable and influenced by a number of risk factors, including hepatitis B virus (HBV), cigarette smoking and alcohol. Due to the complicated pathogenic features of HCC, there is an urgent need to explore valuable prognostic models to increase the possibility of precise prediction and seize therapeutic opportunities for HCC. The European Association for the Study of the Liver (EASL) recommendations recommend alpha-fetoprotein (AFP), vascular endothelial growth factor (VEGF), and Angiopoietin-2 as HCC prognostic indicators (7). Besides, Keratin 19 (K19) and epithelial cellular adhesion molecule (EpCAM) are candidate biomarkers of poor prognosis and invasion in HCC (8). However, the accuracy of prognostic indicators for HCC has proven to be a challenge.
Autophagy, also known as type II programmed cell death, is a well conserved mechanism of self-digestion that employs lysosomes to maintain the equilibrium of cells (9,10). The stability of cellular renewal, homeostasis, and physiological status is crucially dependent on autophagy. The aberrant autophagic level is linked to the pathophysiology of numerous illnesses, including cancer (11). However, the precise function of autophagy in malignancies is unclear and complicated. Various cancer types, tumor microenvironments, and tumor stages have different autophagy pathways (12,13). Given the intricate role that autophagy plays in cancer, more research needs to be conducted into the connections between autophagy and tumors and the underlying biological mechanisms, and the research findings then need to be applied to a well-designed therapeutic strategy to develop a novel approach to cancer therapy. At present, it is too soon to declare with absolute certainty whether autophagy is a friend or foe of cancer (14). Several preclinical studies have recently given evidence on the prognostic roles of autophagy-related gene (ARG) signatures in cancer prognosis (15,16). Based on the promising research results, autophagy modulation, including autophagy inhibitors and autophagy inducers, has been shown to be a potential targeted HCC therapy. Thus, prognostic factors and promising therapies for HCC need to be explored and established.
In this study, we constructed a predictive model for overall survival (OS) prognosis of patients based on ARGs and evaluated the relationship between the expression profiles of the ARGs and clinical outcomes in liver hepatocellular carcinoma (LIHC) patients from The Cancer Genome Atlas (TCGA). Our risk-score approach requires further validation based on additional databases. Our results may lead to a multi-dimensional biomarker technique that is useful at tracking autophagy and gauging the prognosis of LIHC patients. We present the following article in accordance with the TRIPOD reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1130/rc).
Methods
Patient data acquisition
The LIHC training set, which included transcriptome profiling data and corresponding clinical information, was downloaded from TCGA Data Portal (https://tcga-data.nci.nih.gov/tcga/) (17). The validation data set of LIHC patients was downloaded from the International Cancer Genome Consortium (ICGC) database (https://icgc.org/) (18). Table 1 sets out the basic clinical characteristics of the LIHC patients in TCGA database. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Table 1
| Characteristics | Tumor | Normal |
|---|---|---|
| Gender, n | ||
| Male | 236 | 22 |
| Female | 110 | 19 |
| Age (years), n | ||
| Mean | 59 | 61 |
| Median | 61 | 68 |
| Pathologic stage, n | ||
| Stage I | 171 | 18 |
| Stage II | 85 | 10 |
| Stage III | 85 | 12 |
| Stage IV | 5 | 1 |
| Pathologic M, n | ||
| M0 | 263 | 32 |
| M1 | 4 | 1 |
| MX | 79 | 8 |
| Pathologic N, n | ||
| N0 | 253 | 31 |
| N1 | 4 | 1 |
| NX | 89 | 9 |
| Pathologic T, n | ||
| T1 | 173 | 19 |
| T2 | 87 | 10 |
| T3 | 76 | 12 |
| T4 | 10 | 0 |
LIHC, liver hepatocellular carcinoma; TCGA, The Cancer Genome Atlas; M, metastasis; N, node; T, tumor.
Identification of differentially expressed ARGs and enrichment analysis
Differently expressed genes (DEGs) in the LIHC tissue samples and normal solid tissue samples were screened using the “lima” package of R software. The threshold for the DEGs was set as a |log2fold change (FC)| value >1 and a P value <0.05. A total of 1,287 ARGs were acquired from the Human Autophagy Database (HADb; http://autophagy.lu/clustering/index.html) and Autophagy database (http://autophagy.info/). Subsequently, the extracted differentially expressed ARGs underwent further analyses to determine the enriched Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways using the R package “ClusterProfiler” (19).
Construction and validation of the ARG-based prognostic model
To confirm the potential prognostic-related ARGs (P<0.05), a univariate Cox regression analysis was conducted to investigate the association between the expression levels of the ARGs and OS using the “survival” package in R. Subsequently, a multivariable Cox regression analysis was conducted to construct a prognostic model for patient survival and calculate the risk score. The risk score was calculated using the following formula: β × ARG1 expression + β × ARG2 expression + ... + β × ARGn expression (20).
Verification of the ARG-based prognostic signature
We conducted internal validations (in TCGA database) and external validations (in the ICGC database) to assess the predictive performance of the ARG-based prognostic model. All the patients were classified into high- and low-risk groups using the median risk score as the cut-off point. A Kaplan-Meier survival curve analysis was conducted to compare the OS of the patients in the two groups. The accuracy of the ARG-based model was estimated using receiver operating characteristic (ROC) curve and the area under the curve (AUC) values (21). AUC value is lower than 0.5, the identification power of the model is poor. AUC is 0.5–0.7, the model has a low-level identification power. AUC is 0.7–0.9, the model has a certain identification power. AUC is above 0.9, the model has a high-level identification power.
Establishment and validation of nomogram
To assist in clinical procedures, we constructed a prognostic nomogram to assess the probable 1-, 2-, and 3-year OS of LIHC patients (22). The clinicopathological variables included age, gender, and pathological tumor-node-metastasis (TNM) stage. The nomogram and calibration plot were constructed using the “rms” and “survival” package in R software.
Immune infiltration by CIBERSORT analysis
Using 547 different barcode gene expression levels, the CIBERSORT method was used in order to describe the proportion of 22 different immune cells present in tissues. In order to ascertain the percentage of 22 different immune cells present in the LIHC samples, the CIBERSORT algorithm was applied to use. Correlation coefficients of the 22 immune cell were calculated using Pearson correlation analysis (23).
The TIMER database (https://cistrome.shinyapps.io/timer/) was used to estimate the correlations between the gene signatures and tumor-infiltrating immune cells [i.e., B cells, cluster of differentiation (CD)8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells] in TCGA LIHC data set to reveal the immune infiltration of the gene signatures in the cancer. Along with the Spearman’s correlation coefficients and the statistically significant estimates, the correlation module was used to create the expression scatter plots between the gene signatures in LIHC (24).
Statistical analysis
All the statistical analyses were performed using SPSS 24.0 (Chicago, IL, USA) and R software (version 3.5.1). A risk score and two ARG-based nomograms were created using Cox regression coefficients. To evaluate the model’s prediction ability, a time-dependent ROC curve analysis is performed. The log-rank test was used to compare survival curves constructed using the Kaplan-Meier method. The OS was defined as the period between surgery and death or the final follow-up (in days). P value <0.05 was considered statistically significant.
Results
Identification of the differentially expressed ARGs in the LIHC patient tissue samples
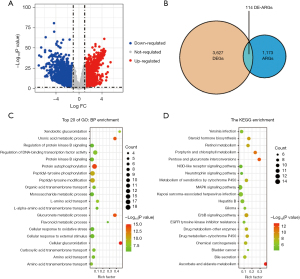
We obtained the transcriptome profiling and clinical prognostic data of the patients in the LIHC cohort from TCGA database (n=346), and we then retrieved 1,287 ARGs from the HADb and Autophagy database (Figure 1A and available oneline: https://cdn.amegroups.cn/static/public/jgo-22-1130-1.xlsx). In total, 114 significant differentially expressed ARGs (46 downregulated and 68 upregulated) were found (Figure 1B and Table S1). We then conducted functional enrichment analyses to further understand the functions and mechanisms of these differentially expressed ARGs. The findings indicated that the 114 ARGs were mostly enriched in protein autophosphorylation, DNA-binding transcription factor activity regulation, and peptidyl-tyrosine phosphorylation. Additionally, the KEGG pathway enrichment analysis revealed that these genes were strongly linked to a number of pathways, including the ascorbate and aldarate metabolism, pentose and glucuronate interconversions, the ErbB signaling system, and the mitogen activated protein kinases signaling network pathway (Figure 1C,1D and Tables S2,S3).
Construction of a prognostic model based on the ARGs
To further isolate the possible prognostic gene from the cohort of ARGs, a univariate Cox regression analysis was conducted. The results revealed that 23 genes were substantially associated with the OS of LIHC patients (P<0.05, Figure 2A). The two ARGs in the training cohort (i.e., PLD1 and SLC36A1) were then identified as independent prognostic markers using multivariate Cox regression (Table S4 and Figure 2B,2C). SLC36A1 was shown to be a risk factor [hazard ratio (HR) >1], while PLD1 was shown to be a protective factor (HR <1). The risk score was calculated as follows: risk score = (−0.441 × PLD1 expression) + (0.522 × SLC36A1 expression).
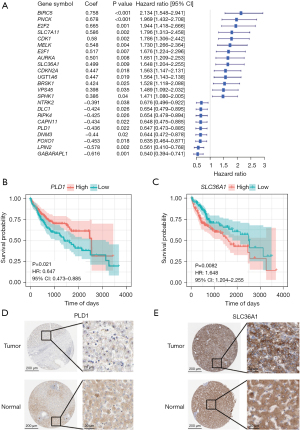
Next, we assessed the levels of PLD1 and SLC36A1 protein expression in the LICH patients. When we examined the immunohistochemistry data from the Human Protein Atlas (https://www.proteinatlas.org/; Figure 2D,2E) (25), we discovered that the expression levels of PLD1 and SLC36A1 in the tumor tissues differed significantly.
Evaluation of the two ARG-based prognostic models
Subsequently, we computed the prognostic risk score for each LIHC patient in the training set and the validation set. Using the median risk score as the cut-off threshold, we divided the patients into two groups; that is, the high-risk group and the low-risk group. The performance of the prognostic model was confirmed using the Kaplan-Meier survival curve and AUC analysis with effective prognosis prediction. Notably, fewer patients in the high-risk group survived than the low-risk group (Figure 3A).
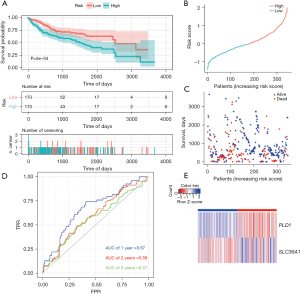
Next, we assessed this model in TCGA cohort using the survival curve and ROC curve results, and a heatmap of gene expression. According to the survival distributions, patients with lower risk scores generally had better survival than those with higher risk scores (Figure 3B,3C). The predictive capability of the two ARG model’s was then assessed using time-dependent ROC curves. The AUCs at 1-, 2-, and 3-year survival were 0.67, 0.59, and 0.57, respectively (Figure 3D). The expression of the two ARGs in the two groups is depicted in a heatmap. In the low-risk group, PLD1 was strongly expressed, while in the high-risk group, SLC36A1 was significantly expressed (Figure 3E).
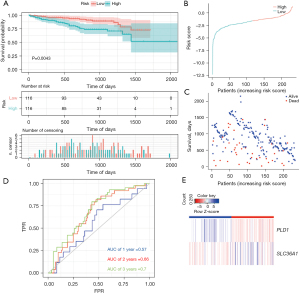
To verify whether the model was reliable, we also used it to examine the external cohort from the ICGC database. Similarly, a Kaplan-Meier curve based on the ROC curve and the log-rank test was generated to show the prognostic value (Figure 4A-4E). The AUCs at 1-, 2-, and 3-year survival were 0.57, 0.66, and 0.7, respectively (Figure 4D). The accuracy of the model in predicting the survival of LIHC patients was supported by both internal and external validations (training set: P=5e-04; validation set: P=0.0043).
Nomogram development for the prediction of prognostic risk
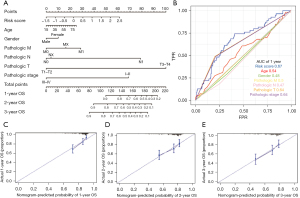
Based on the training set of the LIHC patients, we created a prognostic nomogram to predict OS probability at 1-, 2-, and 3-year using the two ARG classifiers and 6 clinical pathological risk factors. This prognostic model included the following 7 significant independent parameters: risk score, age, gender, M stage, N stage, T stage, and pathologic stage (Figure 5A). The application of the time-dependent ROC curve demonstrated that the nomogram had the highest prognostic accuracy by a sizable margin, indicating that the nomogram prediction model had highly valuable efficacy in predicting early relapse. A time-dependent ROC analysis was done for the 1-year OS to evaluate the prognostic implications of the risk score with other clinical parameters. According to the findings, the risk score had the most predictive impact at this time point, with an AUC value of 0.67 (Figure 5B), which was higher than the AUC values for age (AUC =0.54), gender (AUC =0.48), M stage (AUC =0.5), N stage (AUC =0.47), T stage (AUC =0.64) and pathologic stage (AUC =0.64). No variations from the reference line were observed in the calibration curves of the nomogram, indicating that there was no need for re-calibration [Figure 5C (1-year), Figure 5D (2-year), and Figure 5E (3-year)]. Although the AUC value depicted by the ROC curve did not exceed 0.7, indicating that the prediction impact was relatively inadequate, our findings show that the prognostic signature of ARGs is an independent prognostic factor for HCC patients.
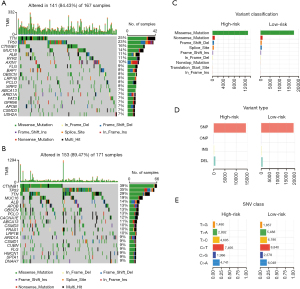
Somatic mutations in different subgroups based on the ARG signature
In addition, typical somatic mutations in individuals in the high- and low-risk groups were investigated using the somatic mutation profiles of the 346 LIHC patients. The “maftools” approach was used to analyze and display the mutation data (26). We observed that the top 5 mutated genes in the high- and low-risk groups were TTN, TP53, CTNNB1, MUC16, and ALB. Waterfall plots were used to indicate mutation data for both high- and low-risk samples of each gene (Figure 6A,6B). Missense mutations constituted the vast majority of mutations in both groups after further categorization based on the numerous comprehensive classifications (Figure 6C). Single nucleotide polymorphism was the most frequent mutation type in both groups (Figure 6D), and the most frequent single nucleotide variant was the C > T transversion (Figure 6E).
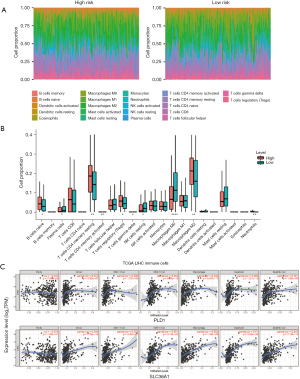
Infiltrating immune cells analyses
We assessed the component proportion of the 22 immune cell subtypes in the LIHC samples using the CIBERSORT algorithm (Figure 7A,7B). The proportions of plasma cells, resting natural killer cells, activated mast cells, and M0 macrophages were relatively lower in the high-risk group than the low-risk group (Wilcoxon test, all P<0.05); however, the high-risk group typically contained higher proportions of naive B cells, resting CD4 memory T cells, regulatory T cells, M2 macrophages, and neutrophils. The associated coefficients between the immune infiltration levels in LIHC patients from TIMER and the two ARGs was then determined using a Pearson correlation analysis. The results revealed that both PLD1 and SLC36A1 expression were significantly correlated with the infiltration levels of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells (Figure 7C).
Discussion
HCC is a serious health concern around the world, particularly in China (27,28). Patients with HCC continue to have poor long-term outcomes even after aggressive resection. The outcomes of HCC patients are closely correlated to the expression of a few genes in the tumor tissue, tumor stage, tumor size, serum indicators, and liver function. To predict the prognosis of HCC patients, several studies have concentrated on the possible relevance of gene signatures based on aberrant messenger ribonucleic acid (29-31). Using data from TCGA as the training set and data from ICGC as the validation set, we combined the gene expression data for HCC in this study.
In recent years, studies on multi-gene prognostic markers for HCC have been reported, such as lncRNAs signature, metabolic-related gene signature, m6A-related gene signature, immune-related gene signature, etc. (32-34). These studies remind researchers that multi-gene combinations may become more reliable prognostic markers for HCC, despite the fact that they have not yet been utilized to guide clinical treatment. Previous research has shown that the development and spread of malignant tumors, inflammatory response, and medication resistance are all highly correlated with autophagy (11). Consequently, the identification of ARG biomarkers could create novel perspectives on the diagnosis of or therapy for different malignancies (35). We sought to examine the predictive value of ARGs that could be used to predict the prognosis of HCC patients. Qin et al. constructed a prognostic model of ARGs for LIHC with partial data sets from TCGA (n=235) (36). However, to build the model in this study, we used the whole TCGA data set (n=346). The two investigations vary in terms of both the differentially expressed ARGs and the models built for them. In addition to developing a predictive model for LIHC, we also examined the function of the ARGs in LIHC in this study.
In this study, we built an ARG prognostic model using high-throughput expression profile data from TCGA and validated this model using data from TCGA and ICGC. First, we identified 114 ARGs that were differentially expressed in the tumor tissues. According to the GO enrichment analysis, these genes were primarily enriched in protein autophosphorylation, DNA-binding transcription factor activity regulation, and peptidyl-tyrosine phosphorylation, which suggests that these DEGs primarily influence autophagy and thus the progression of liver cancer. Ascorbate and aldarate metabolism, pentose and glucuronate interconversions, and the ErbB signaling pathway were the most enriched KEGG pathways. Our findings are in line with those of earlier research that showed dysregulated apoptosis is a prevalent characteristic of liver cancer.
Next, a univariate Cox regression analysis was conducted, and 23 genes were found to be strongly associated with the survival of HCC patients, and two significant genes (i.e., PLD1 and SLC36A1) were ultimately chosen using a multivariate Cox regression analysis for inclusion in the prognostic model (37,38). Based on data from TCGA and the ICGC databases, we divided the patients into high- and low-risk groups and observed that those in the high-risk group had a lower survival rate. After controlling for other clinical factors, we then verified that the risk score generated by the model formula was an independent predictor of survival. Our ROC curve analysis showed that the risk score had a higher predictive value than other clinical criteria.
Evidence of the somatic mutations that drive tumor growth provides a crucial basis for prognostication, therapeutic intervention, and diagnostic practice. In our investigation, we showed that the low-risk score subtype had significantly higher frequencies of TP53 and CTNNB1 mutations than the high-risk score subtype. Previous research has shown that the TP53 mutation leads to the downregulation of the immunotherapeutic response in HCC, and TP53 is one of the most frequently mutated genes in multiple cancer types (39). According to previous reports, the mutation of CTNNB1, which characterizes the immune-excluded phenotype, could serve as a novel indicator for the prediction of immunotherapeutic resistance in HCC (40).
We compared the abundance of the immune cells using the TIMER database and the CIBERSORT algorithm, taking into account the risk signature derived from autophagy, which was significantly correlated with anti-tumor immunity. We discovered that the risk score was positively related to activated immune cells (e.g., naïve B cells, resting memory CD4 T cells, regulatory T cells, M2 macrophages, and neutrophils). These results suggested that immunotherapy could be clinically beneficial for tumor patients with high-risk scores (23,24,41).
In summary, we constructed and validated a prognostic model based on two ARGs. This model could serve as a useful tool in predicting the survival of HCC patients. To our knowledge, the two ARGs included in the prognostic models had not been reported previously, and the differentially expressed ARGs may provide a new perspective for the study of HCC molecular mechanisms. However, it should be noted that our research had some limitations. The AUC value plotted by ROC curve did not exceed 0.7, suggesting that the prediction effect had a low-level identification. Notably, many factors affect liver cancer, among which autophagy genes are only one. Thus, relying solely on ARGs for prediction is not very accurate, and ARGs need to be combined with other factors to ensure comprehensive prediction.
Conclusions
In summary, our study has constructed a robust autophagy-related risk model with two genes (PLD1 and SLC36A1) for OS prediction of HCC. The molecular state of HCC may be reflected in patients’ risk scores. Our study expands the scope of preclinical medicine research and provides new possibilities for predicting patient outcomes and developing individualized treatments for HCC in the future.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1130/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1130/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Liu M, Jiang L, Guan XY. The genetic and epigenetic alterations in human hepatocellular carcinoma: a recent update. Protein Cell 2014;5:673-91. [Crossref] [PubMed]
- Kow AWC. Transplantation versus liver resection in patients with hepatocellular carcinoma. Transl Gastroenterol Hepatol 2019;4:33. [Crossref] [PubMed]
- Doycheva I, Thuluvath PJ. Systemic Therapy for Advanced Hepatocellular Carcinoma: An Update of a Rapidly Evolving Field. J Clin Exp Hepatol 2019;9:588-96. [Crossref] [PubMed]
- Llovet JM, De Baere T, Kulik L, et al. Locoregional therapies in the era of molecular and immune treatments for hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 2021;18:293-313. [Crossref] [PubMed]
- Yang WS, Jiang HY, Liu C, et al. Multi-Omics and Its Clinical Application in Hepatocellular Carcinoma: Current Progress and Future Opportunities. Chin Med Sci J 2021;36:173-86. [PubMed]
- Akhoundova D, Rubin MA. Clinical application of advanced multi-omics tumor profiling: Shaping precision oncology of the future. Cancer Cell 2022;40:920-38. [Crossref] [PubMed]
- European Association for the Study of the Liver. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol 2012;56:908-43. Erratum in: J Hepatol 2012;56:1430. [Crossref] [PubMed]
- Zheng Y, Zhu M, Li M. Effects of alpha-fetoprotein on the occurrence and progression of hepatocellular carcinoma. J Cancer Res Clin Oncol 2020;146:2439-46. [Crossref] [PubMed]
- Patergnani S, Danese A, Bouhamida E, et al. Various Aspects of Calcium Signaling in the Regulation of Apoptosis, Autophagy, Cell Proliferation, and Cancer. Int J Mol Sci 2020;21:8323. [Crossref] [PubMed]
- Yan X, Zhou R, Ma Z. Autophagy-Cell Survival and Death. Adv Exp Med Biol 2019;1206:667-96. [Crossref] [PubMed]
- Klionsky DJ, Petroni G, Amaravadi RK, et al. Autophagy in major human diseases. EMBO J 2021;40:e108863. [Crossref] [PubMed]
- Levine B, Kroemer G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell 2019;176:11-42. [Crossref] [PubMed]
- Kocak M, Ezazi Erdi S, Jorba G, et al. Targeting autophagy in disease: established and new strategies. Autophagy 2022;18:473-95. [Crossref] [PubMed]
- Bhutia SK, Mukhopadhyay S, Sinha N, et al. Autophagy: cancer's friend or foe? Adv Cancer Res 2013;118:61-95. [Crossref] [PubMed]
- Lee PJ, Papachristou GI. New insights into acute pancreatitis. Nat Rev Gastroenterol Hepatol 2019;16:479-96. [Crossref] [PubMed]
- Cao Y, Luo Y, Zou J, et al. Autophagy and its role in gastric cancer. Clin Chim Acta 2019;489:10-20. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013;45:1113-20. [Crossref] [PubMed]
- Zhang J, Bajari R, Andric D, et al. The International Cancer Genome Consortium Data Portal. Nat Biotechnol 2019;37:367-9. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Xue Y, Yan J, Schifano ED. Simultaneous monitoring for regression coefficients and baseline hazard profile in Cox modeling of time-to-event data. Biostatistics 2021;22:756-71. [Crossref] [PubMed]
- Nahm FS. Receiver operating characteristic curve: overview and practical use for clinicians. Korean J Anesthesiol 2022;75:25-36. [Crossref] [PubMed]
- Wang R, Dai W, Gong J, et al. Development of a novel combined nomogram model integrating deep learning-pathomics, radiomics and immunoscore to predict postoperative outcome of colorectal cancer lung metastasis patients. J Hematol Oncol 2022;15:11. [Crossref] [PubMed]
- Newman AM, Steen CB, Liu CL, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol 2019;37:773-82. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Thul PJ, Lindskog C. The human protein atlas: A spatial map of the human proteome. Protein Sci 2018;27:233-44. [Crossref] [PubMed]
- Mayakonda A, Lin DC, Assenov Y, et al. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res 2018;28:1747-56. [Crossref] [PubMed]
- Llovet JM, Kelley RK, Villanueva A, et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2021;7:6. [Crossref] [PubMed]
- Zhao HT, Cai JQ. Chinese expert consensus on neoadjuvant and conversion therapies for hepatocellular carcinoma. World J Gastroenterol 2021;27:8069-80. [Crossref] [PubMed]
- You Y, Hu S. Aberrant expression of the esophageal carcinoma related gene 4 as a prognostic signature for hepatocellular carcinoma. Clin Res Hepatol Gastroenterol 2022;46:101891. [Crossref] [PubMed]
- Cai C, Yang L, Zhou K. 8DEstablishment and validation of a hypoxia-related signature predicting prognosis in hepatocellular carcinoma. BMC Gastroenterol 2021;21:463. [Crossref] [PubMed]
- Zheng S, Xie X, Guo X, et al. Identification of a Pyroptosis-Related Gene Signature for Predicting Overall Survival and Response to Immunotherapy in Hepatocellular Carcinoma. Front Genet 2021;12:789296. [Crossref] [PubMed]
- Zeng H, Liu C, Zhou X, et al. A New Prognostic Strategy Based on four DNA Repair-Associated lncRNAs for Hepatocellular Carcinoma. Comb Chem High Throughput Screen 2022;25:906-18. [Crossref] [PubMed]
- Liu GM, Xie WX, Zhang CY, et al. Identification of a four-gene metabolic signature predicting overall survival for hepatocellular carcinoma. J Cell Physiol 2020;235:1624-36. [Crossref] [PubMed]
- Xia X, Tang P, Liu H, et al. Identification and Validation of an Immune-related Prognostic Signature for Hepatocellular Carcinoma. J Clin Transl Hepatol 2021;9:798-808. [Crossref] [PubMed]
- Lin F, Zhu YT, Qin ZH. Biomarkers of Autophagy. Adv Exp Med Biol 2021;1208:265-87. [Crossref] [PubMed]
- Qin F, Zhang J, Gong J, et al. Identification and Validation of a Prognostic Model Based on Three Autophagy-Related Genes in Hepatocellular Carcinoma. Biomed Res Int 2021;2021:5564040. [Crossref] [PubMed]
- Xiao J, Sun Q, Bei Y, et al. Therapeutic inhibition of phospholipase D1 suppresses hepatocellular carcinoma. Clin Sci (Lond) 2016;130:1125-36. [Crossref] [PubMed]
- Yoshida A, Bu Y, Qie S, et al. SLC36A1-mTORC1 signaling drives acquired resistance to CDK4/6 inhibitors. Sci Adv 2019;5:eaax6352. [Crossref] [PubMed]
- Pinyol R, Torrecilla S, Wang H, et al. Molecular characterisation of hepatocellular carcinoma in patients with non-alcoholic steatohepatitis. J Hepatol 2021;75:865-78. [Crossref] [PubMed]
- Xu C, Xu Z, Zhang Y, et al. β-Catenin signaling in hepatocellular carcinoma. J Clin Invest 2022;132:e154515. [Crossref] [PubMed]
- Llovet JM, Castet F, Heikenwalder M, et al. Immunotherapies for hepatocellular carcinoma. Nat Rev Clin Oncol 2022;19:151-72. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


