
The miRNA125a-5p and miRNA125b-1-5p cluster induces cell invasion by down-regulating DDB2-reduced epithelial-to-mesenchymal transition (EMT) in colorectal cancer
Highlight box
Key findings
• The miRNA125a-5p and miRNA125b-1-5p could reduce the expression of DDB2 by binding to the 3'UTR region, and then regulate the expression levels of EMT markers, leading to the enhanced invasion and metastasis of colorectal cancer cells.
What is known and what is new?
• MiRNA-125 is reportedly aberrantly expressed in colorectal cancer tissue;
• The present study explained the mechanism of miRNA-125 family regulated invasion and migration of colorectal cancer.
What is the implication, and what should change now?
• The miRNA125a-5p and miRNA125b-1-5p might be novel markers of colorectal cancer migration and potential therapeutic targets to treat metastatic colorectal cancer patients.
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumors of the digestive system, and its increasing rates of morbidity and mortality have attracted greater attention in recent decades (1). Approximately 20 percent of CRC cases have reportedly already metastasized at the time of diagnosis, and the 5-year survival rate of metastatic CRC from 1975 to 2009 is only 5.5–12.5% (2). The incidence rate of CRC has increased by 22% from 2000 to 2013 among adults below the age of 50 years. Similarly, the mortality rates increased by 13% in patients under the age of 50 years between 2000 and 2014 (3). However, the invasion and metastasis mechanisms of CRC are poorly understood, which presents a significant problem for clinicians.
MicroRNA (miRNA) is an endogenous small non-coding RNA with a length of about 22 nucleotides, which play crucial roles in important biological processes by regulating multiple proteins expression. Acting at the post-transcriptional level (4-6), miRNA can modulate the expression of as much as 30% of all mammalian protein-encoding genes (7). In some cases, miRNAs are encoded by multiple loci, some of which are organized in tandemly co-transcribed clusters. With the in-depth research on miRNA in recent years, an increasing number of miRNAs have been found to play a key regulatory role in human cancers (8-10), including breast (11), bladder (12), lung (13), and gastric (14) cancers.
MiR-125 is a highly conserved miRNA transcribed from three different clusters in humans, namely, miR-125a on chromosome 19, miR-125b-1 on chromosome 11, and miR-125b-2 on chromosome 21. MiR-125b-1 and miR-125b-2 code for the same mature sequence, and their miRNAs (located in the introns of protein-coding genes) contain putative promoters that regulate transcription independently of their host gene, which differs from most miRNA mechanisms. In addition to paralogs that code for the same mature sequence, there are homologs (e.g., miR-125a) that code for different mature sequences, which contain the same seed region, and therefore, might have similar functions (15). According to previous investigations that employed a next-generation sequencing (NGS)-based profiling study of miRNAs (miRNome), miRNA-125 is present in metastatic CRC, which indicates that there is a close relationship between miR-125 and metastatic CRC (16,17). Additionally, miRNA-125 is positively correlated with hematopoietic malignancy (18). Based on the above findings, we speculate that miRNA-125 might participate in the regulation of CRC metastasis.
In recent decades, numerous studies have demonstrated that the invasion of cancer cells could be induced by several physicochemical factors (19-21); however, their relationship with tumor metastasis is still unknown. Tumor metastasis is a key process of cancer development and occurs via a series of steps, and is the main reason for treatment failure (22). Most CRC patients are only found after metastasis, and the prognosis is poor. Before invasion, cancer cells need to pass through the epithelial-to-mesenchymal transition (EMT) process to obtain migration and invasive properties (23). The essential steps in this process include the loss of cell polarity and achieving a mesenchymal phenotype, which are accompanied by decreased expression of the epithelial marker, E-cadherin, and increased expression of mesenchymal markers, such as N-cadherin and Vimentin (24). However, the potential function and underlying metastasis mechanism of CRC remain unclear, and thus, further investigation is needed (25).
In this study, we aimed to reveal the potential function and mechanism of the miR-125 family in CRC cell lines. We found that miR-125a-5p and miR-125b-1-5p could not only reduce the expression of their targeted gene [damage specific DNA binding protein 2 (DDB2)] but also inhibited the expression of epithelial markers and increased those of mesenchymal cell markers in cancer cells, which was mediated by DDB2. This study might provide helpful information in understanding the function and mechanism of miR-125 family-regulated CRC migration. We present the following article in accordance with the MDAR reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1222/rc).
Methods
Patients and samples
Patients were identified using the electronic records obtained from The First Affiliated Hospital of Nanjing Medical University. Samples were collected from 60 CRC patients from 2012 to 2015, including primary tumors (n=30) and paired metastatic niches (n=30). The methods were performed in accordance with the approved guidelines by the Ethics Committee of The First Affiliated Hospital of Nanjing Medical University (No. 2010-SR-091). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was obtained from all subjects.
Cell lines and cell culture
The SW480 human CRC cell line was obtained from the American Type Culture Collection (ATCC, CCL-228, USA). Cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco, USA) supplemented with 10% fetal bovine serum (FBS, Gibco), 50 mg/mL penicillin (Gibco), 50 mg/mL streptomycin (Gibco), and 1 mM sodium pyruvate. The cells were then cultured at humidified 37 ℃ in 5% carbon dioxide (CO2) and the cell culture medium was replaced every 48 hours.
Transient transfection
Before transfection, SW480 cells were seeded into six-well plates and incubated until 60–70% confluence. The cells were then transfected with miR-125a-5p mimics/sponge, miR-125b-1-5p mimics/sponge, miR-125a-5p and miR-125b-1-5p mimics, miR-125a-5p and miR-125b-1-5p sponge, respectively. Lipo2000 was used for transfection according to the manufacturer’s instructions.
Methyl thiazolyl tetrazolium assay
The SW480 cells were seeded into 96-well plates at a density of 4,000 cells per well and incubated overnight. Forty-eight hours after transfection, the cells were then treated with 20 µL of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide (MTT, Sigma Aldrich, USA) solution [5 mg/mL in phosphate-buffered saline (PBS)] and incubated for 4 h at 37 ℃. Next, the medium was rapidly removed and the MTT crystals were solubilized in 100 µL dimethyl sulfoxide (DMSO). The optical density (OD) volume was determined at 490 nm for each well using a microplate spectrophotometer (BioTek, USA). The data represented the mean ± standard deviation (SD) of at least three independent experiments.
Cell invasion assay
Briefly, the upper surface of the Transwell filter (8.0 µm in pore size) was coated with 40 µg extracellular matrix (ECM) gel before being air-dried overnight at room temperature. Approximately 4×104 cells were added into the upper chamber and cultured overnight. The cells were then transfected with the miRNA mimics/sponge for 6 h, respectively. After 72 h of incubation at 5% CO2 and 37 ℃, the cells invaded the opposite surface of the filter. The filters were fixed with 4% paraformaldehyde and stained with crystal violet, and the cells on the upper surface were then removed completely with a cotton swab. Cell migration was observed under light microscopy (ZEISS, German) at a magnification of ×100. The cells were then lysed using an extraction buffer (Millipore, ECM210, USA) and collected for the detection of OD at 570 nm; the OD value of the treated cells was normalized to the control. Tumor cell invasiveness was defined as the average OD of the cells.
Luciferase reporter assays
The SW480 cells were seeded into 48-well plates at a density of 10,000 cells per well and incubated overnight, then co-transfected with pTA-8×Luc, pRL-TK, and synthetic DDB2 including wild type (WT) or mutant type (MUT). After 48 h, the cells were rinsed with PBS and lysed using a 100 µL passive lysis buffer (PLB). The luciferase activity was assayed using the Dual-Luciferase Reporter Assay System (Promega, USA) and a luminometer. The luciferase activity of each lysate was normalized to the activity of Renilla luciferase driven by the constitutively expressing promoter in the pRL-TK vector.
RNA extraction and quantitative real-time polymerase chain reaction (PCR)
Total RNA was isolated using Trizol (TaKaRa, Japan) reagent according to the manufacturer’s protocol. Total RNA concentrations were determined using a biophotometer (Eppendorf, Germany). Then, 1 µg of purified RNA was reverse transcribed to complementary DNA (cDNA) using the PrimeScript RT Reagent Kit (TaKaRa, Japan). For quantitative real time PCR (qRT-PCR), each reaction was conducted in 96-well plates, with a final volume of 20 µL, including 10 µL SYBR Green PCR Master Mix (TaKaRa, Japan) plus 1 µL primers (2 µM) (Table S1), 1 µL of template DNA, and 7 µL H2O. Each reaction was run in triplicate. Gene expression levels were determined using the comparative threshold cycle 2−ΔΔCt analysis method.
Western blotting
After collection of the cell protein components, a standard Western blotting analysis was employed to investigate the protein expression levels. Briefly, the protein extracts were prepared at the indicated time point using KEYGEN Protein Extraction Kit (KEYGEN, China). Protein lysates were boiled in sodium dodecyl sulfate (SDS) sample loading buffer for 5 minutes, then subjected to 10% SDS polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride (PVDF) membranes (Bio-Rad, USA). The membranes were blocked for 1 hour in 5% milk-Tris-Buffered Saline Tween-20 (TBST) at room temperature and incubated overnight at 4 ℃ with anti-DDB2 (Abcam, ab51017, UK) and anti-GAPDH (Proteintech, 60004-1, China) antibodies, respectively. The membranes were washed three times with TBST and incubated with the appropriate horseradish peroxidase-conjugated secondary antibodies (Proteintech, SA00001-1, China) at room temperature for 2 h. The blots were visualized using enhanced chemiluminescence (Thermo, USA) and analyzed using a scanning densitometer with the molecular analysis software FluorChem M system (Protein Simple, USA).
Statistical analysis
The differences in the invasion rate and protein expression levels between the treated cells and the controls were assessed by one-way analysis of variance (ANOVA) method followed by the standard student t-test analysis. A P value of less than 0.05 was considered statistically significant (P value <0.05). Each experiment was performed at least three times. Computer-based calculations were conducted using SPSS version 20 (SPSS Inc., Chicago, Illinois, USA).
Results
MiRNA-125 family expression characters in primary colorectal tumors and paired metastatic niches
To further explore the relationship between the miRNA-125 family (miR-125a-5p, miR-125b-1-5p, and miR-125b-2-5p) with primary and metastatic tumors, the expression levels of the miRNA-125 family were determined by RT-PCR. The results revealed that miR-125a-5p (Figure 1A) and miR-125b-1-5p (Figure 1B), and not miR-125b-2-5p (Figure 1C), were highly expressed in metastatic tumors compared to primary tumors, indicating that miR-125a-5p and miR-125b-1-5p are correlated with CRC metastasis.

Overexpression of miR-125a-5p and miR-125b-1-5p increases cell invasiveness
A Transwell-filter assay was performed to identify the roles of miR-125a-5p and miR-125b-1-5p in CRC cell invasion. SW480 cells were seeded in a 24-well ECM-coated Transwell chamber and transfected with miR-125a-5p mimics/sponge, miR-125b-1-5p mimics/sponge, miR-125a-5p and miR-125b-1-5p mimics/sponge, respectively. The invaded cells were stained with crystal violet and statistically analyzed by the OD volume.
Compared to the mimics control group (Figure 2A, 1), the miR-125a-5p mimics (Figure 2A, 2) and miR-125b-1-5p mimics (Figure 2A, 3) increased cell invasion ability, respectively, and the miR-125a-5p mimics combined with the miR-125b-1-5p mimics enhanced cell invasion ability, exhibiting a synergistic effect (Figure 2A, 4). The statistically analyzed results revealed that the miR-125a-5p mimics and miR-125b-1-5p mimics increased cell invasiveness by 7.56 and 5.56 folds, respectively, and the combination of miR-125a-5p mimics/miR-125b-1-5p mimics increased cell invasiveness by 12.59 folds (Figure 2B).
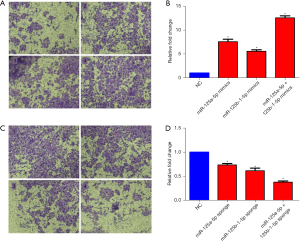
Meanwhile, compared to the sponge inhibitor control group (Figure 2C, 1), the miR-125a-5p sponge (Figure 2C, 2) and miR-125b-1-5p sponge (Figure 2C, 3) reduced the cell invasion ability, respectively, and the miR-125a-5p sponge combined with the miR-125b-1-5p sponge also reduced the cell invasion ability with a synergistic effect (Figure 2C, 4). The statistically analyzed results revealed that the miR-125a-5p sponge and miR-125b-1-5p sponge reduced cell invasiveness by 26.12% and 32.88%, respectively, and the combination of the miR-125a-5p sponge /miR-125b-1-5p sponge reduced cell invasiveness by 46.29% (Figure 2D).
MiRNA-125a-5p and miR-125b-1-5p inhibit DDB2 expression by binding to 3'UTR regions of the DDB2 mRNA
To investigate the potential mechanism of miRNA-125 in regulating colorectal invasion, we primarily predicted DDB2 as the miRNA-125 downstream via crossover analysis in the miRBase, miRDB, and TargetScan databases (Figure 3A). The results showed miR-125a-5p and miR-125b-1-5p bind to the 3'UTR region of DDB2 at positions 221–228 through the “GAGUCCCU” motif.
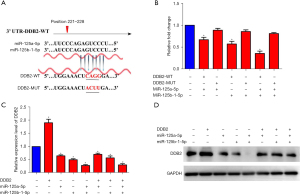
We applied a Dual-Luciferase Reporter Assay to verify the binding sites. We constructed the WT and MUT (CAGG mutated to ACUU) luciferase reporter plasmids of the DDB2 3'UTR region (Figure 3A), then and co-transfected these with miR-125a-5p and miR-125b-1-5p into SW480 cells, respectively. The results showed that compared to the MUT-DDB2 groups, both miR-125a-5p and miR-125b-1-5p could reduce the expression of WT DDB2 with a synergistic effect (Figure 3B), which is consistent with the invasion assay.
We then explored the influence of miR-125a-5p and miR-125b-1-5p on DDB2 expression levels in-depth. A DDB2 expression vector plasmid was constructed for RT-PCR and the Western blotting assay. The qPCR results confirmed DDB2 overexpression and verified that miR-125a-5p and miR-125b-1-5p could reduce the expression of DDB2 in a synergistic manner (Figure 3C), which was confirmed by protein detection (Figure 3D). The gray value of Western binds was also calculated (Figure S1).
MiRNA-125a-5p and miR-125b-1-5p induce CRC cell invasion by inhibiting DDB2
DDB2 was reported to inhibit CRC invasion, and we also detected the DDB2 expression level in primary and metastatic tissues by qRT-PCR. The results showed that DDB2 was significantly down-regulated in the metastatic niches (Figure S2), which is consistent with previously reported findings.
To evaluate the role of DDB2 in miR-125-induced colorectal cell invasion, we transfected the cells with miR-125a-5p and miR-125b-1-5p mimics, respectively, based on DDB2 overexpression. The results revealed that since miR-125a-5p and miR-125b-1-5p increased cell invasion (Figure 4A) by 1.76 and 1.72 folds, respectively (Figure 4B), DDB2 still suppressed miR-125-induced cell invasion compared to the miR-125-overexpressed colorectal cells (Figure 2A).
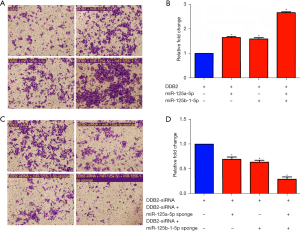
Additionally, we transfected cells with miR-125a-5p and the miR-125b-1-5p sponge, respectively, based on DDB2 depression by small interfering RNA (siRNAs). The results showed that DDB2 suppression covered miR-125 sponge-reduced cell invasion (Figure 4C), which is consistent with Figure 4D. These results indicate that miRNA-125a-5p and miR-125b-1-5p induce CRC cell invasion by inhibiting DDB2.
MiRNA-125a-5p and miR-125b-1-5p activate the CRC cell EMT process via DDB2
It has been reported that DDB2 blocks EMT-mediated metastasis in CRC, so we detected the effects of miR-125a-5p and miR-125b-1-5p on EMT in the SW480 CRC cell line. The qRT-PCR results showed that miR-125a-5p and miR-125b-1-5p increased the expression of the mesenchymal markers Vimentin (Figure 5A) and N-cadherin (Figure 5B), while simultaneously decreasing that of the epithelial marker E-cadherin (Figure 5C).
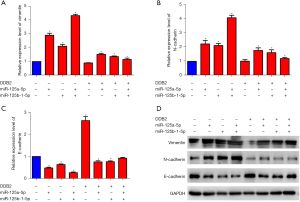
Consistently, the Western blot results demonstrated the same results on protein levels (Figure 5D). DDB2 overexpression suppressed the miR-125a-5p and miR-125b-1-5p-induced EMT process, which is consistent with DDB2-reduced colorectal invasion. These results indicated that miR-125a-5p and miR-125b-1-5p could induce EMT by inhibiting DDB2 expression in CRC cells.
Discussion
CRC is the second most common tumor in the gastrointestinal system. It is associated with high rates of morbidity and mortality, and the incidence of morbidity in young adults is increasing every year, which is a great threat to human health. Unfortunately, the cause of CRC occurrence is still not clear. It is generally believed to be associated with eating habits, with the high consumption of animal fats and proteins as well as low-fiber diets being more likely to cause CRC. Meanwhile, other factors, including genetic susceptibility and pre-cancerous lesions associated with the intestinal tract, are also correlated with CRC (26,27).
The majority of early-stage patients have no obvious symptoms, and most patients are discovered after tumor metastasis. At present, the major prognostic method for CRC involves the TNM staging system (defined by the Tumor, Node, and Metastasis), which determines the treatment decisions and outcomes of patients with CRC. The purpose of staging is to understand the progression of the tumor, guide the proposed treatment plan, and estimate the prognosis of patients (26,28). The liver is the main target of CRC metastasis, and occurs via miRNA regulation; however, the detailed mechanism has not yet been elucidated (29-31).
In this study, we found that miR-125a-5p and miR-125b-1-5p were significantly highly expressed in metastatic CRC tissues (Figure 1), indicating miR-125a-5p and miR-125b-1-5p might be correlated with CRC metastasis. We then performed a Transwell filter assay, which confirmed that the overexpression of miR-125a-5p and miR-125b-1-5p could respectively promote the invasion of CRC cells SW480 (Figure 2). Additionally, the combination of miR-125a-5p and miR-125b-1-5p exhibited a synergistic effect on CRC metastasis.
MiRNAs reportedly affect downstream gene expression by binding to the 3'UTR regions of mRNA (32). To explore the potential mechanism behind miR-125a-5p and miR-125b-1-5p-induced cancer invasion, we predicted the downstream genes by cross-analyzing the miRNA target prediction databases. DDB2 is the target of both miR-125a-5p and miR-125b-1-5p and is depressed in metastatic colorectal tissues (Figure S2). Next, we utilized a luciferase assay to validate whether DDB2 is the direct target of miR-125a-5p and miR-125b-1-5p. The results showed that miR-125a-5p and miR-125b-1-5p could bind to the WT DDB2 but not the MUT, and down-regulate DDB2 dominantly, indicating that miR-125a-5p and miR-125b-1-5p could bind to the 3'UTR regions of DDB2 mRNA. DDB2 has been shown to inhibit colorectal metastasis (33), which is consistent with our findings (Figure 4), illustrating that miR-125a-5p and miR-125b-1-5p increase colorectal cell invasion by down-regulating DDB2.
Moreover, DDB2 is a master suppressor of EMT in CRC. EMT is the process by which epithelial-derived tumor cells transform into stromal cells and acquire the ability to invade and migrate. It exists in multiple physiological and pathological processes in the human body and plays important roles in tissue remodeling and cancer metastasis. Colorectal epithelial cancer cells lose their polarity through EMT and obtain high interstitial phenotypes, such as migration and invasion. To comprehensively elucidate the mechanism of miR-125a-5p and miR-125b-1-5p-induced colorectal invasion, we detected the effect of miR-125a-5p and miR-125b-1-5p on EMT. The results in Figure 5 revealed that miR-125a-5p and miR-125b-1-5p activate the EMT process in CRC cells by inhibiting DDB2. EMT of malignancy cells increased cancer metastatic ability and chemoresistance (34). Increased EMT is also correlated with the formation of cancer stem cells (CSC) (35). Additionally, accumulating evidence indicates that EMT increased CSC population, hence resulting CSC-mediated clinical relapse (36). In our study, we found miRNA125a-5p and miRNA125b-1-5p induced epithelial-mesenchymal transition of CRC, so we proposed miRNA125a-5p and miRNA125b-1-5p could increase CSC properties, including chemoresistance and recurrence. The EMT process contributes to increased CSC population through intra-cellular crosstalk, suggesting it may be reasonable to attenuate EMT process for treatment of malignant CRC (36). It seems a promising strategy of blocking miRNA125a-5p and miRNA125b-1-5p expression to inhibit colorectal EMT and CSC for drug development.
It’s rarely difficult to obtain enough metastatic tissue for detection of miRNAs, so we collected metastatic niches from kinds of parts, including liver, lung, bladder, peritoneum, etc. Next step, we will try to analyze the correlation between miRNA125a-5p and miRNA125b-1-5p expression levels and clinical pathology characters, including frequency of liver metastasis, based on enough cases. Meanwhile, it’s better to verify the effect of miRNA125a-5p and miRNA125b-1-5p on CRC metastasis in vivo, and use multiple methods to assess the effect of miRNA125a-5p and miRNA125b-1-5p on cell invasion and migration.
Conclusions
Our results indicated that miR-125a-5p and miR-125b-1-5p could activate the EMT process by down-regulating DDB2, and thus, enhance the metastatic ability of CRC cells. Our findings preliminarily explored the mechanism of CRC metastasis, which may provide helpful information for future treatment.
Acknowledgments
Funding: This study was financially supported by the National Natural Science Foundation of China (No. 81672788) and the Natural Science Foundation of Jiangsu Province (No. BK20210974).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1222/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1222/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1222/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The methods were performed in accordance with the approved guidelines by the Ethics Committee of The First Affiliated Hospital of Nanjing Medical University (No. 2010-SR-091). Informed consent was obtained from all subjects.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin 2022;72:7-33. [Crossref] [PubMed]
- Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin 2017;67:177-93. [Crossref] [PubMed]
- Siegel RL, Miller KD, Goding Sauer A, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin 2020;70:145-64. [Crossref] [PubMed]
- Wang K. The Ubiquitous Existence of MicroRNA in Body Fluids. Clin Chem 2017;63:784-5. [Crossref] [PubMed]
- Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, et al. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J Cell Physiol 2019;234:5451-65. [Crossref] [PubMed]
- Kabekkodu SP, Shukla V, Varghese VK, et al. Clustered miRNAs and their role in biological functions and diseases. Biol Rev Camb Philos Soc 2018;93:1955-86. [Crossref] [PubMed]
- Vitsios DM, Davis MP, van Dongen S, et al. Large-scale analysis of microRNA expression, epi-transcriptomic features and biogenesis. Nucleic Acids Res 2017;45:1079-90. [Crossref] [PubMed]
- Kara G, Calin GA, Ozpolat B. RNAi-based therapeutics and tumor targeted delivery in cancer. Adv Drug Deliv Rev 2022;182:114113. [Crossref] [PubMed]
- Ali Syeda Z, Langden SSS, Munkhzul C, et al. Regulatory Mechanism of MicroRNA Expression in Cancer. Int J Mol Sci 2020;21:1723. [Crossref] [PubMed]
- Conti I, Varano G, Simioni C, et al. miRNAs as Influencers of Cell-Cell Communication in Tumor Microenvironment. Cells 2020;9:220. [Crossref] [PubMed]
- Saikia M, Paul S, Chakraborty S. Role of microRNA in forming breast carcinoma. Life Sci 2020;259:118256. [Crossref] [PubMed]
- Taheri M, Shirvani-Farsani Z, Ghafouri-Fard S, et al. Expression profile of microRNAs in bladder cancer and their application as biomarkers. Biomed Pharmacother 2020;131:110703. [Crossref] [PubMed]
- Liao Y, Wu X, Wu M, et al. Non-coding RNAs in lung cancer: emerging regulators of angiogenesis. J Transl Med 2022;20:349. [Crossref] [PubMed]
- Pehlevan Özel H, Dinç T, Tiryaki RS, et al. Targeted MicroRNA Profiling in Gastric Cancer with Clinical Assessement. Balkan J Med Genet 2021;24:55-64. [Crossref] [PubMed]
- Wang JK, Wang Z, Li G. MicroRNA-125 in immunity and cancer. Cancer Lett 2019;454:134-45. [Crossref] [PubMed]
- Shirakami Y, Iwashita T, Uemura S, et al. Micro-RNA Analysis of Pancreatic Cyst Fluid for Diagnosing Malignant Transformation of Intraductal Papillary Mucinous Neoplasm by Comparing Intraductal Papillary Mucinous Adenoma and Carcinoma. J Clin Med 2021;10:2249. [Crossref] [PubMed]
- Yin H, Sun Y, Wang X, et al. Progress on the relationship between miR-125 family and tumorigenesis. Exp Cell Res 2015;339:252-60. [Crossref] [PubMed]
- Liu AF, Wang JX, Li FL, et al. Research Progress on miR-125 Family in Malignant Hematologic Diseases-Review. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2017;25:1842-6. [PubMed]
- Mierke CT. The matrix environmental and cell mechanical properties regulate cell migration and contribute to the invasive phenotype of cancer cells. Rep Prog Phys 2019;82:064602. [Crossref] [PubMed]
- Novikov NM, Zolotaryova SY, Gautreau AM, et al. Mutational drivers of cancer cell migration and invasion. Br J Cancer 2021;124:102-14. [Crossref] [PubMed]
- Li X, Bao C, Ma Z, et al. Perfluorooctanoic acid stimulates ovarian cancer cell migration, invasion via ERK/NF-κB/MMP-2/-9 pathway. Toxicol Lett 2018;294:44-50. [Crossref] [PubMed]
- Zeeshan R, Mutahir Z. Cancer metastasis - tricks of the trade. Bosn J Basic Med Sci 2017;17:172-82. [PubMed]
- Brabletz T, Kalluri R, Nieto MA, et al. EMT in cancer. Nat Rev Cancer 2018;18:128-34. [Crossref] [PubMed]
- Cai M, Wang Z, Zhang J, et al. Adam17, a Target of Mir-326, Promotes Emt-Induced Cells Invasion in Lung Adenocarcinoma. Cell Physiol Biochem 2015;36:1175-85. [Crossref] [PubMed]
- Li C, Sun YD, Yu GY, et al. Integrated Omics of Metastatic Colorectal Cancer. Cancer Cell 2020;38:734-747.e9. [Crossref] [PubMed]
- Dekker E, Tanis PJ, Vleugels JLA, et al. Colorectal cancer. Lancet 2019;394:1467-80. [Crossref] [PubMed]
- Qin Y, Havulinna AS, Liu Y, et al. Combined effects of host genetics and diet on human gut microbiota and incident disease in a single population cohort. Nat Genet 2022;54:134-42. [Crossref] [PubMed]
- Biller LH, Schrag D. Diagnosis and Treatment of Metastatic Colorectal Cancer: A Review. JAMA 2021;325:669-85. [Crossref] [PubMed]
- Akbari A, Sedaghat M, Heshmati J, et al. Molecular mechanisms underlying curcumin-mediated microRNA regulation in carcinogenesis; Focused on gastrointestinal cancers. Biomed Pharmacother 2021;141:111849. [Crossref] [PubMed]
- Wang D, Wang X, Si M, et al. Exosome-encapsulated miRNAs contribute to CXCL12/CXCR4-induced liver metastasis of colorectal cancer by enhancing M2 polarization of macrophages. Cancer Lett 2020;474:36-52. [Crossref] [PubMed]
- Tam C, Wong JH, Tsui SKW, et al. LncRNAs with miRNAs in regulation of gastric, liver, and colorectal cancers: updates in recent years. Appl Microbiol Biotechnol 2019;103:4649-77. [Crossref] [PubMed]
- Qiu Y, Yu H, Shi X, et al. microRNA-497 inhibits invasion and metastasis of colorectal cancer cells by targeting vascular endothelial growth factor-A. Cell Prolif 2016;49:69-78. [Crossref] [PubMed]
- Roy N, Bommi PV, Bhat UG, et al. DDB2 suppresses epithelial-to-mesenchymal transition in colon cancer. Cancer Res 2013;73:3771-82. [Crossref] [PubMed]
- Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol 2019;20:69-84. [Crossref] [PubMed]
- Lambert AW, Weinberg RA. Linking EMT programmes to normal and neoplastic epithelial stem cells. Nat Rev Cancer 2021;21:325-38. [Crossref] [PubMed]
- Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol 2017;14:611-29. [Crossref] [PubMed]
(English Language Editor: A. Kassem)


