
The hsa_circ_0007396-miR-767-3p-CHD4 axis is involved in the progression and carcinogenesis of gastric cancer
Highlight box
Key findings
• Cyclic RNA plays a key role in the genesis and development of many human tumors. Our research found that hsa_circ_0007396-miR-767-3p-CHD4 axis is related to the canceration and progression of GC.
What is known and what is new?
• Gastric cancer has a high degree of malignancy and invasion. It is of great significance to reveal the potential molecular mechanism of GC carcinogenesis and progression;
• The low expression of CHD4 is an independent risk factor affecting tumor progression. CHD4 may provide a promising therapeutic target for GC in the future.
What is the implication, and what should change now?
• In the future, relevant
Introduction
Gastric cancer (GC) is one of predominant cancers and the third leading cause of death globally (1). According to statistics, the total number of new cases of GC in the world in 2020 will exceed 1 million, ranking fifth in the number of malignant tumors, and the number of deaths will reach about 769,000, ranking fourth in the number of deaths from malignant tumors (2). GC has a high degree of malignancy, strong invasiveness, and strong heterogeneity. It is difficult to achieve satisfactory therapeutic effects with classical surgery and systemic chemotherapy. According to statistical data, GC has a 10% chance of 5-year survival (3,4). Furthermore, in order to develop new treatment approaches for GC, it is critical to comprehend the fundamental molecular pathways driving the emergence and development of GC carcinogenesis.
Cyclic RNAs are a class of endogenous non coding RNAs with covalent closed loops. Unlike traditional linear RNAs, they are closed RNA molecules formed by reverse splicing. Circular RNA (circRNA) does not have a 5' end cap and a 3' end polyA tail. Through exon cyclization or intron cyclization, 3' and 5' ends are connected to form a complete ring structure, avoiding degradation by nucleic acid exonuclease, so it is more stable and conservative than linear RNA (5). The main biological functions of cyclic RNA include: (I) miRNA sponge function: according to the complementary pairing rule of bases, miRNA inhibits or promotes the translation of mRNA by combining with the untranslated region in the target gene. RNAs can combine with common miRNAs in a competitive way to affect the inhibition of miRNAs on target genes. This type of RNA is called competitive endogenous RNA (ceRNA). CircRNA has a stable structure and contains a large number of miRNA response elements (MREs). It can be used as an efficient ceRNA to combine with miRNA, play the role of miRNA sponge, adsorb miRNA, and effectively inhibit the combination of miRNA and the target gene untranslated region, so as to achieve the regulation of target genes. In addition, some circRNAs can combine with multiple miRNAs to affect the regulation of multiple genes; (II) as a translation template: circRNA lacks the necessary elements for cap dependent translation, such as the 5' cap and polyA tail. Therefore, it is generally considered to have no translation function. However, with the further study of circRNA in recent years, it is found that a few circRNAs contain internal ribosome entry site sequences (IRES), making them capable of translation; (III) regulation of gene expression: Some CircRNAs can combine with transcription factors to regulate gene transcription. exonic circRNA (EciRNA) can specifically bind to some protein molecules in cells, and act as a scaffold to bind to RNA or DNA, providing a platform for the interaction between RNA binding protein (RBP), RNA, and DNA (6). According to recent data, circRNAs are essential in the occurrence and growth of cancer cells. For instance, circRNA cMras controls the miR-567/PTPRG regulatory pathway to slow the development of lung cancer (7). Hepatocellular carcinoma (HCC) development is inhibited by sponging with the help of circRNA CircITCH (has-circ-0001141) (8). In GC, specific cirRNAs have also been shown to contribute to the growth and presence of GC. For instance, the miR-199a-5p/Kloth axis is controlled by circ ITCH, which prevents the spread of GC (9); cyclic RNA_LARP4 inhibits the proliferation and invasion of GC cells by sponging miR-424-5p and regulating the expression of LATS1 (10). The current understanding of circRNAs in GC is still limited, and more investigation is required into the expression patterns and specific mechanisms in GC.
First, we chose differentially expressed circRNAs (DECs) in our study that were differentially expressed between normal gastric tissue and GC tumor tissue. Then, only the significantly up- or down-regulated DECs were screened, and these genes were used as potential GC circRNAs. Following that, putative has-circRNA/miRNA/mRNA regulatory axes associated with GC progression were developed using a variety of in silico analyses, such as expression analysis, miRNA co-prediction, target gene prediction, survival analysis, miRNA-hub gene expression correlation, protein-protein interaction (PPI) network analysis, central gene identification, pathway enrichment analysis, and central gene identification. Finally, immunohistochemical (IHC) staining was used to detect the expression of chromodomain helicase DNA binding protein 4 (CHD4) in a large number of GC tissue samples clinically treated in our hospital to determine the clinicopathological significance. The findings of this study may provide essential guidelines for choosing and creating potential therapies for the cure of GC. We present the following article in accordance with the MDAR reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1218/rc).
Methods
Data analysis using the Gene Expression Omnibus (GEO) database
We selected 2 datasets, GSE83521 and GSE89143, from the GEO database, differential analysis was carried out using Gene Expression Omnibus 2R (GEO2R), and DECs were screened.
circBase analysis
The database circBase (http://www.circbase.org/), which offers a database for investigating known and novel circRNAs in sequencing data, is typically used for assessing circRNA-related studies. Through circBase, data analysis can be performed upstream and downstream of the genome, supporting evidence of their expression can be downloaded, and the datasets can be merged and unified. In our study, the positions of candidate circRNAs were also obtained by Cancer-Specific circRNA Database (CSCD).
CSCD analysis
The CSCD is a database of circRNAs that are particular to cancer, making it easier to research how these circRNAs operate and are regulated. The structural ring diagram of each candidate circRNA was obtained using the CSCD database, and each candidate circRNAs probable MRE locations were predicted.
Circular RNA Interactome (CRI) analysis
To investigate candidate circRNAs binding miRNA, we also developed a network tool for circRNAs and their associated proteins and miRNAs. Expected binding miRNAs of candidate circRNAs can be directly obtained from the CRI database.
StarBase analysis
StarBase (http://starbase.sysu.edu.cn/) is a free and open-source framework for decoding protein-RNA interaction systems, miRNA-non-coding RNA (ncRNA), and miRNA-ceRNA-derived from 108 CLIP-seq datasets produced by 37 studies. In our study, the expression profiles of target miRNAs and genes in GCs were examined using starBase. Additionally, this database was utilized to evaluate the relationship between the expression of miRNAs and their associated target genes in GC. A P value <0.05 was deemed statistically significant.
miRNet analysis
In this study, miRNet may predict probable target genes for miRNAs by connecting miRNAs, targets, and their functions.
Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) analysis
Enrichment analysis and PPI network analysis using the STRING database were performed on the target genes of these putative miRNAs.
Cytoscape software construction
The STRING database provided interacting gene pairs that were again inserted into Cytoscape (https://cytoscape.org/) software. Cell bank calculations then identified the key gene. Additionally, Cytoscape was used to re-enter the circRNA-miRNA or miRNA-hub gene pairings, creating the miRNA-hub gene network or circRNA-miRNA and downloading it.
Clinical data selection
Tissue samples of 100 patients with GC who underwent surgical resection in Sir Run Run Hospital, Nanjing Medical University from January 2015 to September 2017 were selected from the pathology department. There were 30 cases of GC with corresponding paracancer tissue. All these patients were diagnosed with GC only before surgery, and received postoperative adjuvant therapy in accordance with normal time and dose. Follow-up was conducted by outpatient review or telephone follow-up. The outpatient follow-up was conducted once every 3 months in the first year, once every 6 months in the second year and once every year after the second year. Follow-up items: abdominal and pelvic computed tomography (CT) or color ultrasound, chest radiograph, tumor markers, urine routine, liver and kidney function, etc. (additional tests depending on the patient’s condition). Patients ranged in age from 34 to 81 years, with an average age of (60.3±8.4) years. There were 66 male patients and 34 female patients. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Sir Run Run Hospital, Nanjing Medical University (No. 2022-SR-016) and informed consent was taken from all the patients.
Tissue microarray construction
The tissue microarray was prepared by the Pathology Department of Sir Run Run Hospital, Nanjing Medical University. Paraffin blocks of 100 GC tissues were stained with hematoxylin-eosin, and the most typical characteristic markers were selected at the fixation points under the microscope. Each point array contains less than 160 points. Slices three microns thick were cut from the receptor block and transferred to a slide using a tape transfer system for ultraviolet (UV) cross-linking.
IHC
Rabbit polyclonal antibody CHD4 was used. Tissue sections were examined by two experienced pathologists in a double-blind method. The IHC results were scored considering the proportion of positive cells and the intensity of cell staining. IHC results were scored according to the study of Wang et al. (11). Samples with scores below 8 were defined as having low CHD4 expression.
Statistical analysis
Statistical analysis was performed using SPSS 22.0 (IBM) software. The clinicopathologic conditions of the two groups were compared and analyzed by Chi-square test. Survival rate was assessed by Kaplan-Meier method, and significance test was performed by log-rank statistical method. P<0.05 was considered to be statistically significant.
Results
Screening of potential circRNAs in GC
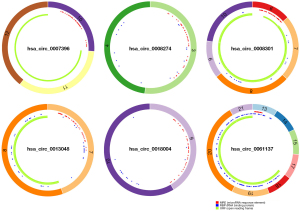
We selected 2 datasets GSE83521 and GSE89143 in the GEO dataset and then performed differential expression analysis on normal gastric tissue and GC by utilizing the GEO2R tool. We found 26 differentially expressed genes (DEGs), as displayed in Figure 1A, Table 1, and Table 2, that were significantly up-regulated or down-regulated in both GSE83521 and GSE89143 compared with normal gastric tissue. Then, we constructed a Venn diagram of the up-regulated and down-regulated DEGs in the 2 datasets, as shown in Figure 1B. In the intersection, we found that there were 0 DEGs that were generally up-regulated, and 0 that were typically down-regulated. There were 8 genes, and we selected 8 DEGs as candidate circRNAs in our study. These 8 potential circRNAs are listed in Table 3, with complete information about them, including the genomic location, circBase ID, and gene probe sequence. In order to further investigate these 8 circRNAs, we also constructed the structural ring diagrams of these genes through the CSCD. We found that only 6 circRNAs had only structural ring diagrams. These circRNAs were hsa_circ_0007396, hsa_circ_0008274, hsa_circ_0008301, hsa_circ_0013048, hsa_circ_0018004, hsa_circ_0061137; therefore, the final candidate circRNAs in GC were thought to be these 6 circRNAs (Figure 2).
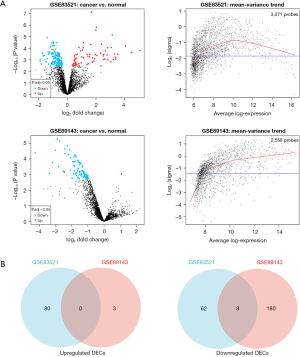
Table 1
| circRNA name | P value | t | B | Log2FC |
|---|---|---|---|---|
| hsa_circRNA_101637 | 0.00000524 | −14.553563 | 4.6751 | −2.896796 |
| hsa_circRNA_103442 | 0.00000727 | −13.783185 | 4.4143 | −2.75262 |
| hsa_circRNA_101592 | 0.00001449 | −12.280053 | 3.8367 | −2.279123 |
| hsa_circRNA_103205 | 0.00002276 | −11.380958 | 3.4407 | −2.430799 |
| hsa_circRNA_102742 | 0.00002625 | −11.110146 | 3.313 | −2.143921 |
| hsa_circRNA_102476 | 0.00002773 | −11.00728 | 3.2634 | −3.016659 |
| hsa_circRNA_103809 | 0.00004063 | −10.316055 | 2.9138 | −4.261945 |
| hsa_circRNA_001826 | 0.00004469 | −10.14982 | 2.8252 | −2.421286 |
| hsa_circRNA_102745 | 0.00005285 | −9.863072 | 2.6681 | −2.201322 |
| hsa_circRNA_102328 | 0.00008704 | −9.052436 | 2.1922 | −2.068563 |
| hsa_circRNA_100146 | 0.0001717 | −8.042424 | 1.5254 | −3.186315 |
| hsa_circRNA_102034 | 0.00031232 | −7.233864 | 0.9234 | −2.814665 |
| hsa_circRNA_102417 | 0.00051793 | −6.603322 | 0.4055 | −2.076447 |
| hsa_circRNA_100269 | 0.00060591 | −6.416857 | 0.2434 | −2.532459 |
| hsa_circRNA_103510 | 0.00063345 | −6.364786 | 0.1974 | −2.463687 |
| hsa_circRNA_001886 | 0.00072733 | −6.204956 | 0.054 | −2.448832 |
| hsa_circRNA_100737 | 0.00082137 | −6.06686 | −0.0726 | −3.382057 |
| hsa_circRNA_000166 | 0.00588838 | −4.115653 | −2.1516 | −3.299404 |
| hsa_circRNA_103219 | 0.00764295 | −3.891177 | −2.4284 | −3.479082 |
| hsa_circRNA_002178 | 0.00765707 | −3.889612 | −2.4304 | −2.107859 |
| hsa_circRNA_000167 | 0.00799667 | −3.852898 | −2.4764 | −2.695141 |
| hsa_circRNA_002172 | 0.00844534 | −3.80696 | −2.5343 | −3.726698 |
| hsa_circRNA_001846 | 0.01172248 | −3.536561 | −2.8816 | −2.771658 |
| hsa_circRNA_000585 | 0.01893475 | −3.156634 | −3.3871 | −2.751842 |
| hsa_circRNA_002144 | 0.02303376 | −3.006011 | −3.5924 | −3.173728 |
| hsa_circRNA_001678 | 0.032568 | −2.745385 | −3.9531 | −2.441521 |
DECs, differentially expressed circRNAs; FC, fold change; circRNAs, circular RNAs.
Table 2
| circRNA name | P value | t | B | Log2FC |
|---|---|---|---|---|
| hsa_circRNA_104589 | 1.56E−06 | 8.502694 | 5.47087 | 2.041879 |
| hsa_circRNA_102592 | 7.09E−06 | 7.36292 | 4.1088 | 3.149847 |
| hsa_circRNA_100267 | 4.49E−05 | 6.111708 | 2.39197 | 2.388223 |
| hsa_circRNA_102191 | 6.34E−05 | 5.893056 | 2.06666 | 4.07073 |
| hsa_circRNA_101471 | 8.74E−05 | 5.693394 | 1.76295 | 3.073238 |
| hsa_circRNA_102584 | 9.17E−05 | 5.66407 | 1.7178 | 2.826348 |
| hsa_circRNA_102614 | 1.54E−04 | 5.349975 | 1.22579 | 5.087543 |
| hsa_circRNA_104915 | 1.70E−04 | 5.288233 | 1.12726 | 2.656947 |
| hsa_circRNA_103122 | 2.36E−04 | 5.09565 | 0.81618 | 4.491761 |
| hsa_circRNA_103053 | 2.67E−04 | 5.022798 | 0.69704 | 2.325002 |
| hsa_circRNA_102561 | 3.18E−04 | 4.921948 | 0.53083 | 3.295099 |
| hsa_circRNA_100984 | 3.34E−04 | 4.892936 | 0.48274 | 3.542264 |
| hsa_circRNA_104947 | 3.45E−04 | 4.8734 | 0.45029 | 2.553155 |
| hsa_circRNA_102815 | 3.84E−04 | 4.812856 | 0.34938 | 2.787683 |
| hsa_circRNA_101909 | 4.31E−04 | 4.746734 | 0.23858 | 2.066083 |
| hsa_circRNA_101309 | 4.58E−04 | 4.711999 | 0.18014 | 2.513364 |
| hsa_circRNA_102777 | 5.93E−04 | 4.565382 | −0.06836 | 3.397041 |
| hsa_circRNA_102615 | 8.55E−04 | 4.360302 | −0.42058 | 3.397793 |
| hsa_circRNA_103594 | 8.55E−04 | 4.36015 | −0.42084 | 2.635859 |
| hsa_circRNA_103559 | 9.56E−04 | 4.298249 | −0.52815 | 3.221738 |
| hsa_circRNA_102571 | 9.59E−04 | 4.296119 | −0.53185 | 2.541839 |
| hsa_circRNA_100385 | 1.01E−03 | 4.267723 | −0.58122 | 2.842232 |
| hsa_circRNA_104807 | 1.51E−03 | 4.046208 | −0.96933 | 3.049633 |
| hsa_circRNA_103783 | 3.40E−03 | 3.60839 | −1.74857 | 2.360019 |
| hsa_circRNA_100269 | 7.59E−03 | −3.18117 | −2.51629 | −2.255969 |
| hsa_circRNA_101308 | 1.32E−02 | 2.888992 | −3.03879 | 2.352548 |
DECs, differentially expressed circRNAs; FC, fold change; circRNAs, circular RNAs.
Table 3
| circRNA name | Gene probe sequence | circBase ID | Location |
|---|---|---|---|
| hsa_circRNA_100571 | TGTTTGCCTGTCAGCAGGTCTCTGTTCTAGGATGTCCCGACCCAGT | hsa_circ_0018004 | chr10:27024168–27024508 |
| hsa_circRNA_103102 | TTGTATAAATGAGCTTACTCCACGGGAACAGCCTCTAGATAATCTG | hsa_circ_0061137 | chr20:61537238–61545758 |
| hsa_circRNA_002117 | AGCAGGGTTGGCGGCGATGTCCGAGGGCCCGGGGCGGCCCCCCTCG | hsa_circ_0000332 | chr11:66626276–66626387 |
| hsa_circRNA_100754 | GCAACCAGAGGCGTGCCGATGCTCTCCGTCCAGCCCAAAGGGAAGC | hsa_circ_0021087 | chr11:8248521–8252051 |
| hsa_circRNA_100737 | TGTGCCCATCACAGGTGTACATCGGTGAGCTCCCGCAGGACTTCCT | hsa_circ_0008301 | chr11:1307231–1317024 |
| hsa_circRNA_100269 | CCCCGATGCCTTCAAAATTATGACTCAAAGATATGAAGATCAATGA | hsa_circ_0013048 | chr1:82302569–82372915 |
| hsa_circRNA_102476 | GCTGGAGCAGGTGAAGCGAGAAATCTTGGTGGAGGTTCTGACCAAA | hsa_circ_0007396 | chr19:17270204–17273932 |
| hsa_circRNA_101287 | CAAGATACAAGATCTCCCCAATAATATGATTTACCAAGTCGCCATT | hsa_circ_0008274 | chr13:96485180–96489456 |
circRNA, circular RNA; GC, gastric cancer.
Analysis and prediction of circRNA-bound miRNAs in GC
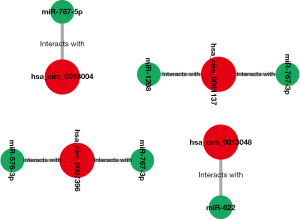
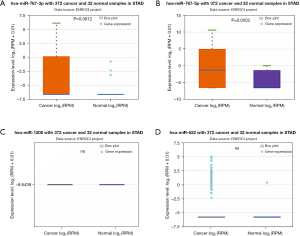
The above 6 final potential circRNA binding miRNAs were predicted using 2 online prediction databases, namely CRI and CSCD. We took the intersection of the miRNAs predicted by these 2 databases, and then obtained the final candidate miRNAs. We found that finally only hsa_circ_0061137, hsa_circ_0018004, hsa_circ_0007396 and hsa_circ_0013048 yielded 6 candidate miRNAs, namely miR-1208, miR-767-3p, miR-767-5p, miR-622, miR-767-3p, and miR-576-3p. For better visualization, we grouped the candidate circRNA-miRNA network composed of these miRNAs and circRNAs, as shown in Figure 3. Subsequently, we used starBase to visualize the expression of these 6 miRNAs in GC, and the results showed that only miR-767-3p and miR-767-5p had statistically key differences (P<0.05), and these 2 miRNAs were significantly different in tumors. The expression in tumor tissue is higher than that in normal tissue (Figure 4). Therefore, we selected miR-767-3p and miR-767-5p as final candidates for circRNA-binding miRNAs.
Analysis and prediction of miRNA selected genes in GC
Then, we built a PPI network in the STRING database using the miRNet database to predict the downstream target genes of miR-767-5p and miR-767-3p. Based on the node degree, we screened out the top 20 hub genes, namely MDM2, SERPINH1, EP300, MMP2, CDK6, LOX, SPARC, EEF2, CASP8, COL5A2, PDGFRB, MCL1, COL3A1, SNW1, FBXW7, NRAS, CDKN1A, CHD4, COL4A2, and COL4A1. The PPI network diagrams of these 20 hub genes are shown in Figure 5. We then performed local network cluster (Table 4), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway (Table 5) and Reactome pathway (Table 6) analyses on these target genes. The KEGG pathway enrichment analysis results included viral carcinogenesis, ubiquitin-mediated proteolysis, miRNAs in cancer, and human papillomavirus infection.

Table 4
| Term ID | Term description | Observed gene count | False discovery rate |
|---|---|---|---|
| CL:16430 | Collagen synthesis and matrix metalloproteinases | 13 | 0.00081 |
| CL:16428 | Mixed, incl. collagen synthesis, and defective b3galtl causes peters-plus syndrome (PPS) | 14 | 0.0038 |
| CL:16432 | Modifying enzymes and collagen biosynthesis | 8 | 0.0143 |
STRING, Search Tool for the Retrieval of Interacting Genes/Proteins.
Table 5
| Term ID | Term description | Observed gene count | False discovery rate |
|---|---|---|---|
| hsa05203 | Viral carcinogenesis | 14 | 0.00042 |
| hsa04120 | Ubiquitin mediated proteolysis | 11 | 0.0018 |
| hsa05206 | MicroRNAs in cancer | 10 | 0.0237 |
| hsa05165 | Human papillomavirus infection | 14 | 0.0429 |
KEGG, Kyoto Encyclopedia of Genes and Genomes.
Table 6
| Term ID | Term description | Observed gene count | False discovery rate |
|---|---|---|---|
| HSA-1474290 | Collagen formation | 9 | 0.0302 |
| HSA-1650814 | Modifying enzymes and collagen biosynthesis | 8 | 0.0302 |
| HSA-1442490 | Collagen degradation | 7 | 0.0324 |
| HSA-1474228 | Degradation of the extracellular matrix | 10 | 0.0324 |
| HSA-1474244 | Extracellular matrix organization | 15 | 0.0324 |
| HSA-2262752 | Cellular responses to stress | 21 | 0.0324 |
| HSA-390471 | Combination of TRiC/CCT with selected proteins during biosynthesis | 6 | 0.0324 |
| HSA-392499 | Metabolism of proteins | 52 | 0.0324 |
| HSA-8948216 | Collagen chain trimerization | 6 | 0.0324 |
| HSA-8953897 | Cellular responses to external stimuli | 22 | 0.0324 |
| HSA-983168 | Antigen processing: proteasome degradation and ubiquitination | 15 | 0.0324 |
| HSA-983169 | Class I MHC mediated antigen presentation and processing | 17 | 0.0324 |
MHC, major histocompatibility complex; TRiC, tailless complex polypeptide 1 ring complex; CCT, chaperonin containing tailless complex polypeptide 1.
Construction of the hsa_circ_0007396-miR-767-3p-CHD4 axis in GC
Next, we evaluated the relationship between the expression of miRNAs and their corresponding target genes in GC through the starBase database (Table 7), and displayed the correlation with a scatter plot (Figure 6). We used Cytoscape to construct a miRNA-hub gene network comprising of the top 20 central genes, miR-767-5p, and miR-767-3p (Figure 7A). According to the correlation size and the prognostic role of target genes in GC, the findings implied that CHD4 may be a significant downstream target of hsa-mir-767-3p in GC (the expression of CHD4 in GC is shown in Figure 7B).
Table 7
| Hub gene | miRNA | R | P value |
|---|---|---|---|
| MDM2 | hsa-mir-767-5p | −0.142 | 5.95E−3 |
| SERPINH1 | hsa-mir-767-5p | 0.126 | 1.48E−2 |
| EP300 | hsa-mir-767-5p | 0.021 | 6.87E−1 |
| MMP2 | hsa-mir-767-5p | −0.105 | 4.35E−2 |
| CDK6 | hsa-mir-767-5p | −0.004 | 9.38E−1 |
| LOX | hsa-mir-767-5p | −0.121 | 1.94E−2 |
| SPARC | hsa-mir-767-5p | −0.140 | 6.74E−3 |
| EEF2 | hsa-mir-767-5p | −0.323 | 1.84E−10 |
| CASP8 | hsa-mir-767-5p | −0.004 | 9.41E−1 |
| COL5A2 | hsa-mir-767-5p | −0.103 | 4.70E−2 |
| PDGFRB | hsa-mir-767-5p | −0.165 | 1.36E−3 |
| MCL1 | hsa-mir-767-5p | −0.063 | 2.22E−1 |
| COL3A1 | hsa-mir-767-5p | −0.148 | 4.18E−3 |
| SNW1 | hsa-mir-767-5p | 0.146 | 4.70E−3 |
| FBXW7 | hsa-mir-767-5p | 0.004 | 9.32E−1 |
| NRAS | hsa-mir-767-5p | 0.041 | 4.26E−1 |
| CDKN1A | hsa-mir-767-3p | −0.081 | 1.18E−1 |
| CHD4 | hsa-mir-767-3p | 0.312 | 7.50E−10 |
| COL4A2 | hsa-mir-767-5p | −0.073 | 1.57E−1 |
| COL4A1 | hsa-mir-767-5p | −0.059 | 2.55E−1 |
miRNA, microRNA; GC, gastric cancer; CHD4, chromodomain helicase DNA binding protein 4.
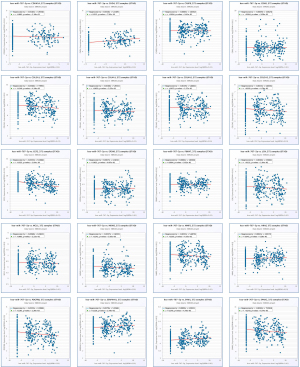
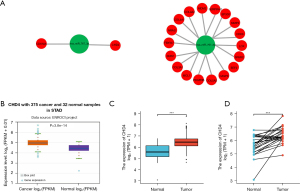
In the above study, hsa_circ_0007396 is a down-regulated circRNA, miR-767-3p is positively correlated with CHD4, and CHD4 is expressed more strongly in GC cancer tissues than in nearby tissues (Figure 7C,7D). Therefore, we believe that the hsa_circ_0007396-miR-767-3p-CHD4 axis might be crucial in the development of GC cancer.
Correlation between CHD4 expression and clinicopathological parameters in patients with GC
Among the 100 GC patients included, 45 cases (45.0%) in the group with low CHD4 expression and 55 cases (55.0%) in the group with high CHD4 expression. There were no statistically significant differences between the two groups in gender, age, weight loss, CEA, serum albumin, tumor size, tumor differentiation, tumor site, TNM stage, chemotherapy, distant metastasis, and nerve invasion (all P>0.05) (Table 8).
Table 8
| Variable | Total, n (%) | CHD4 low expression, n (%) | CHD4 high expression, n (%) | P |
|---|---|---|---|---|
| Gender | 0.520 | |||
| Male | 66 (66.0) | 28 (42.4) | 38 (57.6) | |
| Female | 34 (34.0) | 17 (50.0) | 17 (50.0) | |
| Age (years) | 0.092 | |||
| <64 | 55 (55.0) | 24 (43.6) | 31 (56.4) | |
| ≥64 | 45 (45.0) | 21 (46.7) | 24 (53.3) | |
| Loss of body mass (kg) | 0.105 | |||
| <5 | 56 (56.0) | 26 (46.4) | 30 (53.6) | |
| ≥5 | 44 (44.0) | 19 (43.2) | 25 (56.8) | |
| CEA (ng/mL) | 3.778 | |||
| ≤5 | 56 (56.0) | 30 (53.6) | 26 (46.4) | |
| >5 | 44 (44.0) | 15 (34.1) | 29 (65.9) | |
| Serum albumin (g/L) | 1.010 | |||
| <35 | 80 (80.0) | 34 (42.5) | 46 (57.5) | |
| ≥35 | 20 (20.0) | 11 (55.0) | 9 (45.0) | |
| Tumor size (cm) | 5.533 | |||
| <3 | 63 (63.0) | 34 (54.0) | 29 (46.0) | |
| ≥3 | 37 (37.0) | 11 (29.7) | 26 (70.3) | |
| Differentiation of tumor | 4.302 | |||
| High to middle differentiation | 53 (53.0) | 29 (54.7) | 24 (45.3) | |
| Low differentiation | 47 (47.0) | 16 (34.0) | 31 (66.0) | |
| Site of tumor | 0.253 | |||
| High-medium | 36 (36.0) | 15 (41.7) | 21 (58.3) | |
| Low | 64 (64.0) | 30 (46.9) | 34 (53.1) | |
| TNM stage | 10.725 | |||
| I–II | 32 (32.0) | 22 (68.8) | 10 (31.2) | |
| III–IV | 68 (68.0) | 23 (33.8) | 45 (66.2) | |
| Depth of infiltration | 0.049 | |||
| In the early (Tis + T1) | 21 (21.0) | 9 (42.9) | 12 (57.1) | |
| Stage of progression (T2–T4) | 79 (79.0) | 36 (45.6) | 43 (54.4) | |
| Chemotherapy | 3.093 | |||
| Yes | 64 (64.0) | 33 (51.6) | 31 (48.4) | |
| No | 36 (36.0) | 12 (33.3) | 24 (66.7) | |
| Distant transfer | 0.088 | |||
| Yes | 8 (8.0) | 4 (50.0) | 4 (50.0) | |
| No | 92 (92.0) | 41 (44.6) | 51 (55.4) | |
| Lymph node metastasis | 0.042 | |||
| Yes | 4 (4.0) | 2 (50.0) | 2 (50.0) | |
| No | 96 (96.0) | 43 (44.8) | 53 (55.2) | |
| Invasion of nerve | 1.584 | |||
| Yes | 21 (21.0) | 12 (57.1) | 9 (42.9) | |
| No | 79 (79.0) | 33 (41.8) | 46 (58.2) |
CEA, carcinoembryonic antigen; TNM, tumor/lymph nodes/metastasis; CHD4, chromodomain helicase DNA binding protein 4.
Comparison of OS and PFS in patients with high or low CHD4 expression
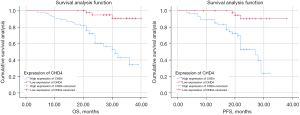
We analyzed the relationship between CHD4 expression and patients’ higher overall survival (OS) and progression-free survival (PFS). The results showed that patients with low CHD4 expression had significantly OS and PFS than patients with high CHD4 expression (Figure 8).
Log-rank univariate analysis and Cox regression analysis
Univariate analysis showed that weight loss, tumor size, tumor differentiation, TNM stage, chemotherapy or not, and low CHD4 expression were factors affecting patients’ OS (P<0.05). Cox multivariate analysis showed that low CHD4 expression was an independent risk factor for tumor progression (P=0.001), and tumor differentiation and chemotherapy also had a certain impact on the progression of GC (all P<0.05). Indicated that increased CHD4 expression predicts poor prognosis in patients with GC (Tables 9,10).
Table 9
| Variables | HR | 95% CI | P |
|---|---|---|---|
| Gender | |||
| Male | 1.581 | 0.730–3.420 | 0.245 |
| Female | 1.000 | – | – |
| Age (years) | |||
| <64 | 1.119 | 0.526–2.383 | 0.770 |
| ≥64 | 1.000 | – | – |
| Loss of body mass (kg) | |||
| <5 | 2.276 | 1.054–4.912 | 0.036 |
| ≥5 | 1.000 | – | – |
| CEA (ng/mL) | |||
| ≤5 | 1.564 | 0.731–3.345 | 0.249 |
| >5 | 1.000 | – | – |
| Serum albumin (g/L) | |||
| <35 | 1.000 | ||
| ≥35 | 0.034 | 0.001–1.693 | 0.090 |
| Tumor size (cm) | |||
| <3 | 3.125 | 1.443–6.768 | 0.004 |
| ≥3 | 1.000 | – | – |
| Differentiation of tumor | |||
| High to middle differentiation | 3.782 | 1.631–8.770 | 0.002 |
| Low differentiation | 1.000 | – | – |
| Site of tumor | |||
| High-medium | 1.000 | ||
| Low | 0.661 | 0.308–1.416 | 0.287 |
| TNM stage | |||
| I–II | 13.777 | 1.867–101.66 | 0.010 |
| III–IV | 1.000 | – | – |
| Depth of infiltration | |||
| In the early (Tis + T1) | 1.000 | ||
| Stage of progression (T2–T4) | 0.474 | 0.212–1.059 | 0.069 |
| Chemotherapy | |||
| Yes | 13.569 | 4.680–39.344 | <0.001 |
| No | 1.000 | – | – |
| Distant transfer | |||
| Yes | 2.713 | 0.366–20.113 | 0.329 |
| No | 1.000 | – | – |
| Lymph node metastasis | |||
| Yes | 1.457 | 0.196–10.831 | 0.713 |
| No | 1.000 | – | – |
| Invasion of nerve | |||
| Yes | 31.659 | 0.735–1364.330 | 0.072 |
| No | 1.000 | – | – |
| CHD4 | |||
| Low expression | 9.255 | 2.774–30.884 | <0.001 |
| High expression | 1.000 | – | – |
OS, overall survival; CEA, carcinoembryonic antigen; TNM, tumor/lymph nodes/metastasis; HR, hazard Ratio; CI, confidence interval, CHD4, chromodomain helicase DNA binding protein 4.
Table 10
| Variable | HR | 95% CI | P |
|---|---|---|---|
| Differentiation of tumor | |||
| High to middle differentiation | 1.000 | ||
| Low differentiation | 0.338 | 0.143–0.799 | 0.014 |
| Chemotherapy | |||
| Yes | 1.000 | ||
| No | 0.088 | 0.03–0.254 | <0.001 |
| CHD4 | |||
| Low expression | 1.000 | ||
| High expression | 0.130 | 0.038–0.439 | 0.001 |
HR, hazard Ratio; CI, confidence interval, CHD4, chromodomain helicase DNA binding protein 4.
Discussion
GC is one of the most common types of human cancer worldwide. Although the general outlook for GC patients is favorable, more studies are required to better understand the specific molecular pathways behind GC pathogenesis. To reveal the expression and mechanism of circRNAs in GC, differential expression analysis was performed on the GSE83521 and GSE89143 datasets in the GEO database using GEO2R. Then, we learned and constructed the structural loop diagram of these genes through the CSCD database, and finally we identified 6 candidate circRNAs. Some of these circRNAs, such as hsa_circ_100571, which was expressed at a higher level on day 1 following liver transplantation than before transplantation, have been shown to be directly associated with biological processes, such as the development and progression of human tumors (P<0.01), and these levels decreased after transplantation. Therefore, it might be utilized as a new biomarker for the identification of primary liver cancer (12); by deactivating the PI3K/Akt axis, up-regulated hsa_circRNA_100269 prevents the development and spread of GC (13).
According to Salmena et al. (14), extended ncRNAs, transcribed pseudogenes, and mRNAs can “talk” to one another utilizing MREs as the alphabet of a brand-new language. The action of ceRNA creates a massive regulatory network in the transcriptome, considerably enhancing the functional genetic information in the human genome and being significant in pathological circumstances like cancer. Current findings have also experimentally confirmed that circRNAs, such as circ100876, can control gene expression via sponge-shared miRNAs, influencing the course of GC. CircRNAs, such as circRNA_100876 sponge miR-136 to promote GC proliferation and metastasis by increasing the expression of MIEN1 (15); circRNA_100782 promotes GC metastasis and proliferation by adsorbing miR-574-3p in a sponge form and downregulating the tumor suppressor gene Rb (16); the circRNA circLMO7 behaves like a sponge for miRNA-30a-3p to boost gastric tumor growth via the WNT2/β-catenin pathway (17); circNRIP1 behaves like a miRNA-149-5p sponge to boost the advancement of GC via the AKT1/mTOR pathway (18). The binding miRNAs of these candidate circRNAs were predicted through the combination of two databases, namely CSCD and CRI.
In conclusion, we obtained 6 candidate miRNAs for these candidate circRNAs, among which only miR-767-3p and miR-767-5p had statistically significant differences, and the expression of these 2 miRNAs in tumor tissues was higher than normal in the organization. The ceRNA hypothesis states that circRNAs and their bound miRNAs are negatively correlated. MiR-767-5p and miR-767-3p have been found to play essential roles in the biological events of other human tumors, for example, miR-767-3p prevents the progression and movement of lung adenocarcinoma cells by regulating CLDN18, even though their potential mechanistic functions in GC have not been explored (19); circRNA hsa_circ_0000673 boosts HCC malignancy by reducing SET-targeting miR-767-3p (20); circRNA circ_0000190 regulates the miR-767-5p/MAPK4 pathway inhibits the growth of multiple myeloma (21); circNOL10 inhibits breast cancer progression through the regulation of SOCS2/JAK/STAT signaling through cavernous miR-767-5p (22).
Using an extensive target gene prediction database (i.e., miRNet), we identified the target genes for miR-767-5p and miR-767-3p to examine the downstream action mechanism. It is generally accepted that genes generally exert their functions through interactions. Moreover, the STRING database was used to analyze the PPI network utilizing these projected target genes. The top 20 core genes in the PPI network were then screened, and a miRNA-hub gene network was created. According to the magnitude of the correlation among them and the prognostic role of target genes in GC, we found that only CHD4 may be a key downstream target of hsa-mir-767-3p in GC. CHD4 is a member of the CHD-like (also known as chromodomain helicase DNA-binding protein) protein family and a member of the SWI2/SNF2-related ATPase superfamily. Among them, CHD4 and CHD5 together form the second subgroup of this family according to the conservation of the encoded protein sequence (23,24). According to the current findings, several biological processes, including the occurrence and growth of various malignancies, have been linked to CHD4. For example, CHD4 increases cancer stemness and epithelial to mesenchymal transition (EMT) in papillary thyroid carcinoma (PTC) cells and predicts aggression in PTC patients (25). In human HCC, the CHD4/NuRD complex controls complement gene expression and is correlated with CD8 T cell infiltration (26); CHD4 mediates non-small cell lung cancer migration and proliferation by RhoA/ROCK pathway via regulating PHF5A (27). Chang et al. (28) found that CHD4 affected the cell motility and drug sensitivity of colorectal cancer (CRC) cells in vitro. In animal models, the depletion of CHD4 affects the growth of CRC tumors, and the combination of histone deacetylase 1 (HDAC1) inhibitors and platinum drugs inhibits the expression of CHD4 and increases the cytotoxicity of platinum drugs. The expression of CHD4 is a feasible biomarker to predict metastatic CRC patients, and has the potential to become a drug development target; Wang et al. proposed that the overexpression of CHD4 is an independent biomarker of poor prognosis in rectal cancer patients receiving concurrent radiotherapy and chemotherapy (11). Meanwhile, it has been revealed that in GC F5A, CHD4 functions as an oncogene (29), which is also consistent with our findings in the starBase database and The Cancer Genome Atlas (TCGA) database. These studies, along with the outcomes of earlier research, suggest that hsa_circ_0007396-miR-767-3p may affect the expression and function of CHD4, allowing it to operate as a mediator of cancer proliferation and oncogenicity. Finally, we verified the clinical data of patients with GC in our hospital, and our IHC results also confirmed that the expression of CHD4 in GC tissues was higher than that in paracancer tissues. CHD4 tissue expression, tumor differentiation and chemotherapy are independent prognostic factors for GC. Kaplan-Meier survival analysis showed that patients with high CHD4 expression showed lower OS and PFS than those with low CHD4 expression.
Conclusions
In summary, this study, combined with some bioinformatics analysis, identified the potential pathway hsa_circ_0007396-miR-767-3p-CHD4 axis in GC, and validated and analyzed clinical data. CHD4 may be an effective target for the development of GC therapy. Therefore, in the future, primary research and clinical studies will need to confirm the current findings.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1218/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1218/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1218/coif). YW, PW and YZ are from Guangzhou Weimi Biotechnology Co., Ltd. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Sir Run Run Hospital, Nanjing Medical University (No. 2022-SR-016) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Abrams T, Hess LM, Zhu YE, et al. Predictors of heterogeneity in the first-line treatment of patients with advanced/metastatic gastric cancer in the U.S. Gastric Cancer 2018;21:738-44. [Crossref] [PubMed]
- Caba L, Florea L, Gug C, et al. Circular RNA-Is the Circle Perfect? Biomolecules 2021;11:1755. [Crossref] [PubMed]
- Luo J, Liu H, Luan S, et al. Guidance of circular RNAs to proteins' behavior as binding partners. Cell Mol Life Sci 2019;76:4233-43. [Crossref] [PubMed]
- Yu C, Tian F, Liu J, et al. Circular RNA cMras inhibits lung adenocarcinoma progression via modulating miR-567/PTPRG regulatory pathway. Cell Prolif 2019;52:e12610. [Crossref] [PubMed]
- Guo X, Wang Z, Deng X, et al. Circular RNA CircITCH (has-circ-0001141) suppresses hepatocellular carcinoma (HCC) progression by sponging miR-184. Cell Cycle 2022;21:1557-77. [Crossref] [PubMed]
- Wang Y, Wang H, Zheng R, et al. Circular RNA ITCH suppresses metastasis of gastric cancer via regulating miR-199a-5p/Klotho axis. Cell Cycle 2021;20:522-36. [Crossref] [PubMed]
- Zhang J, Liu H, Hou L, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer 2017;16:151. [Crossref] [PubMed]
- Wang HC, Chou CL, Yang CC, et al. Over-Expression of CHD4 Is an Independent Biomarker of Poor Prognosis in Patients with Rectal Cancers Receiving Concurrent Chemoradiotherapy. Int J Mol Sci 2019;20:4087. [Crossref] [PubMed]
- Sui W, Gan Q, Liu F, et al. The differentially expressed circular ribonucleic acids of primary hepatic carcinoma following liver transplantation as new diagnostic biomarkers for primary hepatic carcinoma. Tumour Biol 2018;40:1010428318766928. [Crossref] [PubMed]
- Wang Z, Liu C. Upregulated hsa_circRNA_100269 inhibits the growth and metastasis of gastric cancer through inactivating PI3K/Akt axis. PLoS One 2021;16:e0250603. [Crossref] [PubMed]
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353-8. [Crossref] [PubMed]
- Yu X, Xiao W, Song H, et al. CircRNA_100876 sponges miR-136 to promote proliferation and metastasis of gastric cancer by upregulating MIEN1 expression. Gene 2020;748:144678. [Crossref] [PubMed]
- Xin D, Xin Z. CircRNA_100782 promotes roliferation and metastasis of gastric cancer by downregulating tumor suppressor gene Rb by adsorbing miR-574-3p in a sponge form. Eur Rev Med Pharmacol Sci 2020;24:8845-54. [PubMed]
- Cao J, Zhang X, Xu P, et al. Circular RNA circLMO7 acts as a microRNA-30a-3p sponge to promote gastric cancer progression via the WNT2/β-catenin pathway. J Exp Clin Cancer Res 2021;40:6. [Crossref] [PubMed]
- Zhang X, Wang S, Wang H, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer 2019;18:20. [Crossref] [PubMed]
- Wan YL, Dai HJ, Liu W, et al. miR-767-3p Inhibits Growth and Migration of Lung Adenocarcinoma Cells by Regulating CLDN18. Oncol Res 2018;26:637-44. [Crossref] [PubMed]
- Jiang W, Wen D, Gong L, et al. Circular RNA hsa_circ_0000673 promotes hepatocellular carcinoma malignance by decreasing miR-767-3p targeting SET. Biochem Biophys Res Commun 2018;500:211-6. [Crossref] [PubMed]
- Feng Y, Zhang L, Wu J, et al. CircRNA circ_0000190 inhibits the progression of multiple myeloma through modulating miR-767-5p/MAPK4 pathway. J Exp Clin Cancer Res 2019;38:54. [Crossref] [PubMed]
- Wang F, Wang X, Li J, et al. CircNOL10 suppresses breast cancer progression by sponging miR-767-5p to regulate SOCS2/JAK/STAT signaling. J Biomed Sci 2021;28:4. [Crossref] [PubMed]
- Hall JA, Georgel PT. CHD proteins: a diverse family with strong ties. Biochem Cell Biol 2007;85:463-76. [Crossref] [PubMed]
- Mills AA. The Chromodomain Helicase DNA-Binding Chromatin Remodelers: Family Traits that Protect from and Promote Cancer. Cold Spring Harb Perspect Med 2017;7:a026450. [Crossref] [PubMed]
- Pratheeshkumar P, Siraj AK, Divya SP, et al. CHD4 Predicts Aggressiveness in PTC Patients and Promotes Cancer Stemness and EMT in PTC Cells. Int J Mol Sci 2021;22:504. [Crossref] [PubMed]
- Shao S, Cao H, Wang Z, et al. CHD4/NuRD complex regulates complement gene expression and correlates with CD8 T cell infiltration in human hepatocellular carcinoma. Clin Epigenetics 2020;12:31. [Crossref] [PubMed]
- Xu N, Liu F, Wu S, et al. CHD4 mediates proliferation and migration of non-small cell lung cancer via the RhoA/ROCK pathway by regulating PHF5A. BMC Cancer 2020;20:262. [Crossref] [PubMed]
- Chang CL, Huang CR, Chang SJ, et al. CHD4 as an important mediator in regulating the malignant behaviors of colorectal cancer. Int J Biol Sci 2021;17:1660-70. [Crossref] [PubMed]
- Kim MS, Chung NG, Kang MR, et al. Genetic and expressional alterations of CHD genes in gastric and colorectal cancers. Histopathology 2011;58:660-8. [Crossref] [PubMed]
(English Language Editor: J. Jones)


