
Identification of a potential competing endogenous RNA (ceRNA) network in gastric adenocarcinoma
Highlight box
Key findings
• We constructed a ceRNA network of gastric adenocarcinoma.
What is known and what is new?
• ceRNA hypothesis suggested that a large-scale regulatory network was formed in the transcriptome by crosstalk between coding RNAs and non-coding RNAs via MREs (microRNA response elements, MREs).
• Combined with the difference of prognosis and expression, screening and verification were conducted layer by layer.
What is the implication, and what should change now?
• This study was a bioinformatics analysis based on the ceRNA hypothesis, further exploration and verification of RNAs from the network are needed.
Introduction
Gastric adenocarcinoma, as a multifactorial disease, is currently the fifth most common type of cancer and the third most common cause of cancer death globally (1). Risk factors for the condition include Helicobacter pylori infection, age, high salt intake, and diets low in fruit and vegetables (2). Though some causal factors of gastric adenocarcinoma have been identified recently, its etiology and pathogenesis remain obscure (3). Moreover, the lack of differential symptoms results in a low diagnostic rate for advanced gastric adenocarcinoma (4). Therefore, more effective screening methods are required to diagnose gastric adenocarcinoma. A great deal of research has shown that non-coding RNA is closely related to the occurrence and development of cancers (5-7). Competing endogenous RNA (ceRNA) is transcripts that can be mutually regulated at the post-transcriptional level by competing for shared microRNAs (miRNAs). The ceRNA network links the function of protein-coding messenger RNA (mRNA) with non-coding RNA (8,9). Bioinformatics analysis has been widely used in basic research. Therefore, this study aimed to identify the hub genes and construct a ceRNA network for gastric adenocarcinoma using bioinformatics analyses. We present the following article in accordance with the REMARK reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1201/rc).
Methods
Microarray data
Microarray data related to gastric adenocarcinoma were retrieved according to the criteria of “Homo sapiens”, “expression was analyzed through array”, “gastric cancer group and control group were included” and “sample size ≥30”. The datasets GSE54129 (Affymetrix GPL570 platform), GSE118916 (Affymetrix GPL15207 platform), and GSE13861 (Affymetrix GPL6884 platform) were obtained from the Gene Expression Omnibus (GEO) website (https://www.ncbi.nlm.nih.gov/geo/). The Limma software package (version: 3.40.2) of R software was used to explore the expression of mRNA. The GES54129 dataset contained 111 gastric adenocarcinoma samples and 21 noncancerous samples. GSE118916 contained 15 gastric adenocarcinoma samples and 15 noncancerous samples, and GSE13861 contained 60 gastric adenocarcinoma samples and 19 noncancerous samples. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Identification of differentially expressed genes (DEGs)
A GEO2R analysis was provided directly by the GEO project. A P value <0.01, a log fold change (FC) value ≥1, or a logFC value ≤−1 were defined as the thresholds for the DEGs. The overlapping DEGs were obtained by Venn diagrams.
Construction of the protein-protein interaction (PPI) network and selection of hub genes
The PPI network of the DEGs was constructed using the STRING database (http://string-db.org/). Combined scores with interactions >0.4 were considered statistically significant. The PPI networks were visualized by Cytoscape software (version 3.7.1). CytoHubba, a plugin of Cytoscape, sorted the nodes into a network according to the network characteristics. Using the degree calculation, we defined the top 20 hub genes.
Enrichment analysis of DEGs and hub genes
We conducted Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses on the DEGs and hub genes by using the Database for Annotation, Visualization and Integrated Discovery (DAVID) version 6.8 (https://david.ncifcrf.gov/home.jsp). A P value <0.05 was considered statistically significant.
Association of hub gene expressions with the survival of gastric adenocarcinoma patients
The expression and prognostic value of the hub genes were analyzed using the Gene Expression Profiling Interactive Analysis (GEPIA) website and the Kaplan-Meier plotter online tool, and the results were applied to the search for key genes. Then the differential expression of key genes was verified using the Oncomine database analysis (http://www.oncomine.org).
Construction of the miRNA subnet
The correlation between key genes and miRNA expression was analyzed by the miRNet 2.0 online database (https://www.mirnet.ca/miRNet/home.xhtml). Meanwhile, the Kaplan-Meier plotter was used to analyze the prognostic value of the predicted miRNAs on overall survival.
Prediction of upstream long noncoding RNAs (lncRNAs)
The miRNet database was used to predict the upstream lncRNAs of the miRNAs. The Kaplan-Meier plotter was used to evaluate the prognostic value of the predicted lncRNAs on overall survival. The differential expressions in gastric adenocarcinoma and normal tissues were analyzed by the Encyclopedia of RNA Interactomes (ENCORI) online dataset.
Construction of the lncRNA-miRNA-mRNA network
ENCORI 3.0 (http://starbase.sysu.edu.cn/) was used to evaluate the mRNA-lncRNA, miRNA-mRNA, and mRNA-lncRNA pairs of common expression in gastric adenocarcinoma.
Results
Identification of DEGs in gastric adenocarcinoma
We identified 1,155 genes in the GSE118916 dataset, 598 genes in the GSE13816 dataset, and 1,751 genes in the GSE54129 dataset as DEGs by standardization of the microarray results. The overlap among the three datasets is shown as a Venn diagram (Figure 1 and Table 1) with 182 genes included. Two genes were excluded because of inconsistent regulation. The DEGs comprised 53 upregulated and 127 downregulated genes in gastric adenocarcinoma tissues.
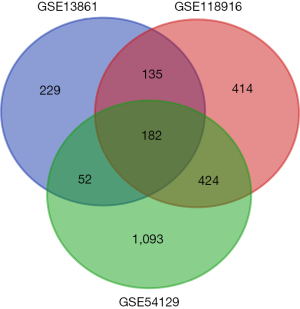
Table 1
| DEGs | Gene |
|---|---|
| Upregulated | CDH3, TGFBI, COL4A2, FAM83D, PLA2G2A, CEMIP, HOXB7, CPXM1, SULF2, SULF1, THBS1, CHRDL2, CLDN3, TNFRSF11B, SFRP4, THBS4,I FITM3, RARRES1, BGN, FNDC1, TMEM158, COL5A2, THBS2, COL6A3, COL1A2, MMP7, PLA2G7, OLFML2B, IGF2BP3, COMP, COL10A1, SERPINH1, COL3A1, FAP, COL4A1, SPP1, MMP3, LY6E, THY1, NID2, CTHRC1, FOXC1, CXCL1, PLAU, COL1A1, PTGS2, CXCL8, SNX10, CLDN1, TIMP1, CST1, COL8A1, CDH11 |
| Downregulated | UBL3 ,POU2AF1 ,CLDN18, ANKRD22, SMIM24, ADTRP, IGFBP2, IRX3, AADAC, ADGRG2, CYP2C9, ESRRG, GUCA2B, TFF1, KCNJ16, PIGR, RNASE1, CBLIF, AMPD1, GKN2, CYP2C18, CAPN8, HPGD, PLAC8, C6ORF58, TMPRSS2, SULT2A1, C1ORF116, FBP2, CA2, BCAS1, GPT, PSCA, RDH12, TRNP1, MAL, SCIN, SULT1C2, MLPH, KRT20, CYP3A5, PIK3C2G, S100P, ITPKA, LRRC31, FOXA1, SCGB2A1, SLC28A2, DHRS7, MUCL3, CPA2, ATP4A, SSTR1, LIPF, NRG4, CTSE, ST6GALNAC1, STX19, KCNE2, CXCL17, ARL14, CWH43, ALDH1A1, FCGBP, FA2H, SCNN1B, SELENBP1, ARHGEF37, NQO1, ATP4B, NOSTRIN, LYPD6B, ANG, TCN1, REG1A, ALDH3A1, SST,T MEM171, KIAA1324, JCHAIN, ADRB2, PRSS8, AKR1C3, FAM174B, GATA5, PGC, CHGA, LTF, AKR7A3, CA9, CA4, TFF2, VILL, GPAT3, REG3A, FAM3B, MRAP2, TFCP2L1, MAOA, ADAM28, VSIG2, CAPN13, AGR2, SOSTDC1, EPN3, CYSTM1, PTGR1, TNFRSF17, GC, SPINK1, ADH7, DDX60, GPT2, TENT5C, ANXA10, KLK11, ADH1C, CAPN9, SPTSSB, AKR1B10, MFSD4A, PLAAT2,S TOX2, VSIG1, GKN1, TMED6, DNER |
DEGs, differentially expressed genes.
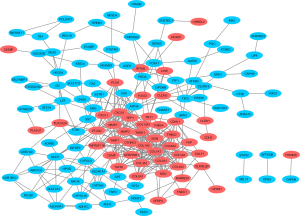
The PPI network analysis.
A PPI network was constructed using 130 nodes and 390 edges to identify the key genes, as shown in Figure 2.
The selection of hub genes
According to the CytoHubba degree method, 20 hub genes were identified: ATP4A, BGN, COL1A1, COL1A2, COL3A1, COL4A1, COL4A2, COL5A2, COL6A3, COL10A1, CXCL1, CXCL8, MMP3, PTGS, SERPINH1, SPP1, THBS1, THBS2, TIMP1, and MMP7.
The enrichment analysis of DEGs
The DAVID website was used to perform the GO annotation and KEGG pathway enrichment analysis to ascertain the biological functions of the DEGs. Extracellular matrix (ECM) organization, digestion, collagen fibril organization, collagen catabolic process, and skeletal system development were the top five biological processes (BP) of the DEGs. The cellular composition (CC) of the DEGs mainly included extracellular space, extracellular exosome, extracellular region, ECM, and endoplasmic reticulum lumen. ECM structural constituent, oxidoreductase activity, ECM binding, platelet-derived growth factor binding, and calcium ion binding were the molecular functions (MF) of the DEGs. ECM-receptor interaction, protein digestion and absorption, focal adhesion, chemical carcinogenesis, and retinol metabolism were the main KEGG functional pathways of the DEGs (Figure 3).
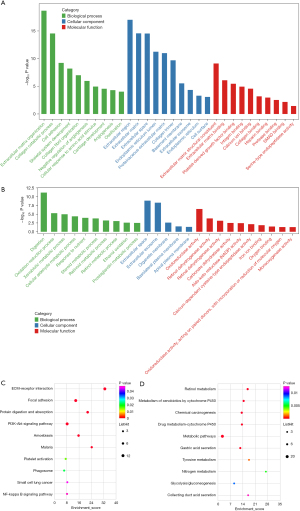
The enrichment analysis of the hub genes
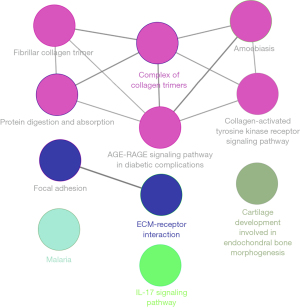
The enrichment of the GO and KEGG pathways was analyzed to explore the function of the hub genes. The most significant KEGG pathways of the hub genes were ECM-receptor interaction, focal adhesion, protein digestion and absorption, the PI3K-Akt signaling pathway, and amoebiasis. The significant GO annotations were collagen catabolic process, endoplasmic reticulum lumen, ECM organization, extracellular region, ECM, collagen trimer, and ECM structural constituent (Table 2). The hub gene enrichment analysis results were visualized by ClueGo of Cytoscape (Figure 4).
Table 2
| ID | Term | Count | P value | FDR | Gene |
|---|---|---|---|---|---|
| GO:0030574 | Collagen catabolic process | 10 | 8.43E-18 | 2.12E-15 | COL1A1, COL3A1, MMP7, COL1A2, COL4A2, COL4A1, COL5A2, MMP3, COL10A1, COL6A3 |
| GO:0005788 | Endoplasmic reticulum lumen | 11 | 1.12E-15 | 4.16E-14 | COL1A1, COL3A1, COL1A2, COL4A2, COL4A1, COL5A2, SERPINH1, COL10A1, COL6A3, PTGS2, THBS1 |
| GO:0030198 | Extracellular matrix organization | 11 | 3.11E-15 | 3.91E-13 | COL1A1, COL3A1, COL1A2, COL4A2, COL4A1, COL5A2, SPP1, BGN, COL10A1, COL6A3, THBS1 |
| GO:0005576 | Extracellular region | 17 | 9.58E-15 | 1.77E-13 | CXCL8, MMP7, MMP3, BGN, CXCL1, THBS2, THBS1, COL1A1, COL3A1, COL1A2, COL4A2, COL4A1, COL5A2, SPP1, COL6A3, COL10A1, TIMP1 |
| GO:0031012 | Extracellular matrix | 11 | 8.91E-14 | 1.10E-12 | COL1A1, COL3A1, MMP7, COL1A2, COL4A2, COL4A1, COL5A2, BGN, COL6A3, THBS2, THBS1 |
| hsa04512 | ECM-receptor interaction | 10 | 1.21E-13 | 5.47E-12 | COL1A1, COL3A1, COL1A2, COL4A2, COL4A1, COL5A2, SPP1, COL6A3, THBS2, THBS1 |
| GO:0005581 | Collagen trimer | 8 | 3.18E-12 | 2.94E-11 | COL1A1, COL3A1, COL1A2, COL5A2, SERPINH1, COL10A1, COL6A3, TIMP1 |
| GO:0005201 | Extracellular matrix structural constituent | 7 | 8.10E-11 | 4.13E-09 | COL1A1, COL3A1, COL1A2, COL4A2, COL4A1, COL5A2, BGN |
| hsa04510 | Focal adhesion | 10 | 3.22E-10 | 7.24E-09 | COL1A1, COL3A1, COL1A2, COL4A2, COL4A1, COL5A2, SPP1, COL6A3, THBS2, THBS1 |
| hsa04974 | Protein digestion and absorption | 8 | 7.72E-10 | 1.16E-08 | COL1A1, COL3A1, COL1A2, COL4A2, COL4A1, COL5A2, COL10A1, COL6A3 |
| GO:0005615 | Extracellular space | 13 | 7.84E-10 | 5.80E-09 | CXCL8, MMP7, MMP3, CXCL1, THBS1, COL1A1, ATP4A, COL3A1, COL1A2, SERPINH1, SPP1, COL6A3, TIMP1 |
| GO:0005578 | Proteinaceous extracellular matrix | 8 | 5.96E-09 | 3.68E-08 | MMP7, COL1A2, COL5A2, MMP3, BGN, COL10A1, COL6A3, TIMP1 |
| hsa04151 | PI3K-Akt signaling pathway | 10 | 3.08E-08 | 3.47E-07 | COL1A1, COL3A1, COL1A2, COL4A2, COL4A1, COL5A2, SPP1, COL6A3, THBS2, THBS1 |
| GO:0030199 | Collagen fibril organization | 5 | 9.39E-08 | 7.86E-06 | COL1A1, COL3A1, COL1A2, COL5A2, SERPINH1 |
| hsa05146 | Amoebiasis | 7 | 1.25E-07 | 1.13E-06 | COL1A1, COL3A1, CXCL8, COL1A2, COL4A2, COL4A1, COL5A2 |
| GO:0048407 | Platelet-derived growth factor binding | 4 | 1.98E-07 | 5.06E-06 | COL1A1, COL3A1, COL1A2, COL4A1 |
| GO:0071230 | Cellular response to amino acid stimulus | 5 | 2.02E-07 | 1.27E-05 | COL1A1, COL3A1, COL1A2, COL4A1, COL5A2 |
| GO:0001501 | Skeletal system development | 5 | 1.49E-05 | 7.50E-04 | COL1A1, COL3A1, COL1A2, COL5A2, COL10A1 |
| GO:0022617 | Extracellular matrix disassembly | 4 | 8.19E-05 | 0.003428113 | MMP7, MMP3, SPP1, TIMP1 |
| GO:0050840 | Extracellular matrix binding | 3 | 3.84E-04 | 0.006525144 | SPP1, BGN, THBS1 |
GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; FDR, false discovery rate.
The mRNA expression and survival analysis
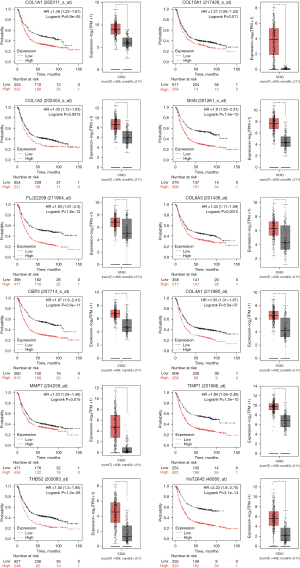
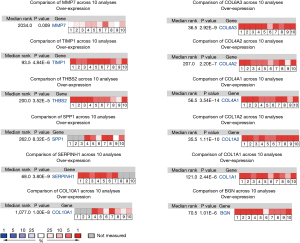
Sources from the Genotype-Tissue Expression (GTEx) and The Cancer Genome Atlas (TCGA) databases were used to analyze the expression levels of the hub genes. The gastric adenocarcinoma samples possessed higher expressions of BGN, COL1A1, COL1A2, COL3A1, COL4A1, COL4A2, COL5A2, COL6A3, COL10A1, CXCL1, CXCL8, MMP3, SERPINH1, SPP1, THBS2, TIMP1, and MMP7 compared with normal samples. Only ATP4A was downregulated in gastric adenocarcinoma tissues. Moreover, ATP4A, COL10A1, BGN, COL1A1, COL1A2, COL4A1, COL4A2, COL5A2, SERPINH1, SPP1, THBS2, TIMP1, and MMP7 were related to poor overall survival in gastric adenocarcinoma, as identified by the Kaplan-Meier plotter. By comprehensive differential expression and survival analysis, we identified COL10A1, BGN, COL1A1, COL1A2, COL4A1, COL4A2, COL6A3, SERPINH1, SPP1, THBS2, TIMP1, and MMP7 as key genes (Figure 5). The differential expression of the key genes was verified by the Oncomine database analysis (http://www.oncomine.org) (Figure 6).
Prediction and validation of the miRNAs
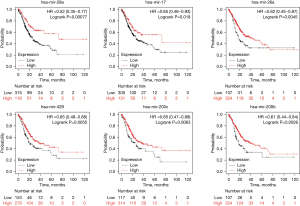
The upstream miRNAs of the key genes were predicted using miRNet 2.0. Our results showed that 18 miRNAs (hsa-mir-29a-3p, hsa-mir-29b-3p, hsa-mir-200b-3p, hsa-mir-9-5p, hsa-mir-125a-5p, hsa-mir-200c-3p, hsa-mir-29c-3p, hsa-mir-429, hsa-mir-503-5p, hsa-mir-26a-5p, hsa-mir-10b-5p, hsa-mir-17-5p, hsa-mir-20a-5p, hsa-mir-22-3p, hsa-mir-497-5p, hsa-mir-224-5p, hsa-mir-29b, and hsa-mir-29c) could potentially regulate the key genes. Based on the classical inverse relationship theory, it is assumed that upstream miRNAs may exert a positive prognostic effect. Using the Kaplan-Meier plotter tool, we found six of the 18 miRNAs (hsa-mir-29a-3p, hsa-mir-200b-3p, hsa-mir-200c-3p, hsa-mir-429, hsa-mir-26a-5p, and hsa-mir-17-5p) were associated with prognosis in gastric adenocarcinoma patients, as shown in Figure 7. The six miRNAs were defined as key miRNAs. The miRNA-mRNA network was obtained by Cytoscape software (Figure 8).
Prediction of lncRNAs
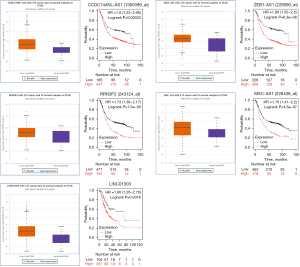
The prediction of upstream lncRNAs of the six miRNAs was conducted by miRNet. Strong evidence for the potential binding of 157 lncRNAs to the six miRNAs was found. A negative correlation between lncRNAs and miRNAs was supported by the hypothesis that lncRNAs could competitively bind to miRNAs. We identified 40 lncRNAs associated with a poor prognosis for gastric adenocarcinoma patients that had a high expression in gastric adenocarcinoma tissues: ARRDC1-AS1, CCDC144NL-AS1, DLGAP1-AS1, DLX6-AS1, GABPB1-AS1, GAS5, HCG18, LINC00638, LINC00852, LINC00879, LINC00943, LINC00997, LINC01111, LINC01270, LINC01553, LINC-PINT, MIAT, MIRLET7BHG, MMP25-AS1, NNT-AS1, PSMD6-AS2, PVT1, SNHG16, SNHG17, STK4-AS1, THUMPD3-AS1, WASIR2, MATN1-AS1, PTPRG-AS1, LINC00174, MSC-AS1, ZEB1-AS1, NUTM2B-AS1, KCNQ1OT1, HELLPAR, MAPKAPK5-AS1, LINC01303, RRN3P2, MCM3AP-AS1, and LINC00894). The 40 lncRNAs were defined as key lncRNAs (Figure 9).
Construction of the ceRNA regulatory network
Based on the previous prediction, there were 18 mRNA-miRNA pairs, 86 miRNA-lncRNA pairs, and 54 mRNA-lncRNA pairs. According to the ceRNA hypothesis, miRNAs have an opposite co-expression relationship with mRNAs and lncRNAs, whereas lncRNAs have a positive co-expression relationship with mRNAs. We assessed the correlation between all RNA interaction pairs using the ENCORI database, and found that 13 out of 18 mRNA-miRNA pairs, 18 out of 86 miRNA-lncRNA pairs, and 18 out of 54 mRNA-lncRNA pairs were consistent with the ceRNA rule (Table 3). Finally, 7 mRNAs (BGN, COL1A1, COL4A1, COL4A2, COL6A3, SERPINH1, MMP7) , 3 miRNAs (hsa-mir-29a-3p, hsa-mir-200b-3p, hsa-mir-429) and 5 lncRNAs (MSC-AS1, ZEB1-AS1, LINC01303, RRN3P2, CCDC144NL-AS1) constructed 24 mRNA-miRNA-lncRNA triple regulatory ceRNA network as a potential regulatory network for gastric carcinoma (Table 4). Cytoscape was used to visualize the ceRNet (Figure 10).
Table 3
| mRNA | miRNA | lncRNA | R | P |
|---|---|---|---|---|
| MMP7 | hsa-mir-429 | −0.125 | 1.56E-02 | |
| BGN | hsa-mir-429 | −0.344 | 9.18E-12 | |
| COL4A1 | hsa-mir-429 | −0.277 | 5.78E-08 | |
| COL1A1 | hsa-mir-29a-3p | −0.159 | 2.04E-03 | |
| COL6A3 | hsa-mir-29a-3p | −0.239 | 3.17E-06 | |
| COL1A2 | hsa-mir-26a-5p | −0.128 | 1.38E-02 | |
| SERPINH1 | hsa-mir-200b-3p | −0.118 | 2.23E-02 | |
| COL4A2 | hsa-mir-200b-3p | −0.410 | 1.62E-16 | |
| MMP7 | hsa-mir-200b-3p | −0.107 | 3.91E-02 | |
| BGN | hsa-mir-200b-3p | −0.366 | 3.16E-13 | |
| COL4A2 | hsa-mir-17-5p | −0.349 | 4.55E-12 | |
| COL1A2 | hsa-mir-17-5p | −0.292 | 9.41E-09 | |
| COL4A1 | hsa-mir-17-5p | −0.192 | 1.91E-04 | |
| hsa-mir-29a-3p | CCDC144NL-AS1 | −0.253 | 7.47E-07 | |
| hsa-mir-29a-3p | HCG18 | −0.274 | 8.25E-08 | |
| hsa-mir-29a-3p | LINC00638 | −0.173 | 7.87E-04 | |
| hsa-mir-17-5p | MIRLET7BHG | −0.112 | 3.08E-02 | |
| hsa-mir-17-5p | PSMD6-AS2 | −0.128 | 1.32E-02 | |
| hsa-mir-200b-3p | MSC-AS1 | −0.428 | 5.46E-18 | |
| hsa-mir-200c-3p | MSC-AS1 | −0.494 | 2.73E-24 | |
| hsa-mir-429 | MSC-AS1 | −0.432 | 2.39E-18 | |
| hsa-mir-200b-3p | ZEB1-AS1 | −0.205 | 6.87E-05 | |
| hsa-mir-200c-3p | ZEB1-AS1 | −0.223 | 1.40E-05 | |
| hsa-mir-429 | ZEB1-AS1 | −0.237 | 3.92E-06 | |
| hsa-mir-29a-3p | KCNQ1OT1 | −0.157 | 2.39E-03 | |
| hsa-mir-200b-3p | LINC01303 | −0.257 | 5.25E-07 | |
| hsa-mir-200c-3p | LINC01303 | −0.227 | 9.89E-06 | |
| hsa-mir-429 | LINC01303 | −0.184 | 3.55E-04 | |
| hsa-mir-200b-3p | RRN3P2 | −0.219 | 2.12E-05 | |
| hsa-mir-200c-3p | RRN3P2 | −0.297 | 5.44E-09 | |
| hsa-mir-429 | RRN3P2 | −0.303 | 2.44E-09 | |
| BGN | MSC-AS1 | 0.741 | 2.05E-66 | |
| BGN | ZEB1-AS1 | 0.252 | 7.67E-07 | |
| BGN | LINC01303 | 0.323 | 1.54E-10 | |
| BGN | RRN3P2 | 0.197 | 1.23E-04 | |
| COL1A1 | CCDC144NL-AS1 | 0.281 | 3.29E-08 | |
| COL4A1 | MSC-AS1 | 0.483 | 2.54E-23 | |
| COL4A1 | LINC01303 | 0.273 | 7.63E-08 | |
| COL4A1 | RRN3P2 | 0.181 | 4.39E-04 | |
| COL4A2 | LINC00997 | 0.139 | 7.19E-03 | |
| COL4A2 | MSC-AS1 | 0.546 | 1.50E-30 | |
| COL4A2 | LINC01303 | 0.242 | 2.11E-06 | |
| COL4A2 | RRN3P2 | 0.225 | 1.05E-05 | |
| COL6A3 | CCDC144NL-AS1 | 0.277 | 4.92E-08 | |
| MMP7 | MSC-AS1 | 0.152 | 3.09E-03 | |
| MMP7 | LINC01303 | 0.165 | 1.36E-03 | |
| MMP7 | RRN3P2 | 0.16 | 1.92E-03 | |
| SERPINH1 | MSC-AS1 | 0.295 | 6.01E-09 | |
| SERPINH1 | LINC01303 | 0.159 | 2.02E-03 |
mRNAs, messenger RNA; miRNA, microRNA; lncRNAs, long noncoding RNAs.
Table 4
| LncRNA | miRNA | mRNA |
|---|---|---|
| CCDC144NL-AS1 | hsa-mir-29a-3p | COL1A1 |
| CCDC144NL-AS1 | hsa-mir-29a-3p | COL6A3 |
| MSC-AS1 | hsa-mir-429 | BGN |
| LINC01303 | hsa-mir-429 | BGN |
| RRN3P2 | hsa-mir-429 | BGN |
| ZEB1-AS1 | hsa-mir-429 | BGN |
| LINC01303 | hsa-mir-429 | MMP7 |
| MSC-AS1 | hsa-mir-429 | MMP7 |
| RRN3P2 | hsa-mir-429 | MMP7 |
| LINC01303 | hsa-mir-429 | COL4A1 |
| MSC-AS1 | hsa-mir-429 | COL4A1 |
| RRN3P2 | hsa-mir-429 | COL4A1 |
| MSC-AS1 | hsa-mir-200b-3p | BGN |
| ZEB1-AS1 | hsa-mir-200b-3p | BGN |
| LINC01303 | hsa-mir-200b-3p | BGN |
| RRN3P2 | hsa-mir-200b-3p | BGN |
| LINC01303 | hsa-mir-200b-3p | MMP7 |
| MSC-AS1 | hsa-mir-200b-3p | MMP7 |
| RRN3P2 | hsa-mir-200b-3p | MMP7 |
| LINC01303 | hsa-mir-200b-3p | COL4A2 |
| MSC-AS1 | hsa-mir-200b-3p | COL4A2 |
| RRN3P2 | hsa-mir-200b-3p | COL4A2 |
| LINC01303 | hsa-mir-200b-3p | SERPINH1 |
| MSC-AS1 | hsa-mir-200b-3p | SERPINH1 |
ceRNAs, competing endogenous RNAs; lncRNAs, long noncoding RNAs; miRNA, microRNA; mRNAs, messenger RNA.
Discussion
Gastric adenocarcinoma has a high incidence and poor prognosis. Its mechanism, suggested by some studies as a latent correlation between the disease and ncRNAs (15) or miRNAs (16), requires further in-depth exploration. Additionally, the effects of ceRNAs in the context of genetic regulatory networks are still poorly understood. Identification and analysis of ceRNA network may reveal the potential pathogenesis of gastric adenocarcinoma at the molecule level and help facilitate gastric adenocarcinoma diagnosis and treatment.
In our study, the intersection of DEGs was taken from three GEO datasets (GSE54129, GSE118916, and GSE13861) for analysis, with a total of 180 DEGs identified. Results of the KEGG pathway enrichment analysis demonstrated that ECM-receptor interaction, protein digestion, focal adhesion, and absorption were the most significant factors. Moreover, the GO enrichment analysis identified ECM organization, endoplasmic reticulum lumen, collagen catabolic process, and extracellular region as the most significant factors. In addition, we identified the top 20 hub genes by constructing a PPI, including 19 upregulated genes and one downregulated gene. Interestingly, similar results for hub genes were obtained by the GO and KEGG enrichments, which indicated a close association between gastric adenocarcinoma and adhesion and ECM-related molecular mechanisms (17). Notably, the disruption of intercellular adhesion and the degradation of ECM have been found to promote the overactivated invasiveness and metastases of cancer cells (18).
Recently, a growing body of research has demonstrated the complex networks between miRNAs and lncRNAs. Based on the ceRNA hypothesis (9), lncRNAs act as important post-transcriptional regulators of downstream gene expressions through miRNA mediation and take part in the pathological processes of cancer. For instance, Wang et al. reported that miR-1290 promotes the proliferation and invasion of chordoma (19), Liu et al. found that the GATA3-AS1/miR-30b-5p/Tex10 axis regulates tumorigenesis of pancreatic cancer (20), Zhou et al. identified that SNHG4/miR-590-3p/CDK1 modulates the colorectal cancer cell cycle and cell proliferation (21), and Chen et al. reported that HIF1A-AS2/miR-30a-5p/SOX4 accelerates the malignant phenotypes of renal carcinoma (22). Furthermore, ceRNA networks were also reported to play an important role in gastric adenocarcinoma. Yang et al. found that LINC01133/miR-106a-3p/APC exerts an impact on gastric adenocarcinoma migration (23), Zhang et al. demonstrated that lncRNA MT1JP competitively binds to miR-92a-3p and functions as a ceRNA to regulate FBXW7 in gastric adenocarcinoma (24), and Chen et al. identified that the LINC01234/miR-204-5p/CBFB axis is crucial in the tumorigenesis of gastric adenocarcinoma (25).
Encouragingly, we constructed 24 mRNA-miRNA-lncRNA triple regulatory networks by stepwise reverse prediction from mRNA to lncRNA. Different from other prediction construction networks, we adjusted layer by layer and verified layer by layer based on ceRNA hypothesis and combined with prognostic and expression differences. Each RNA in the network is of essential prognostic value in gastric adenocarcinoma. Some studies proved that miRNA-429 contributes to the development of multiple cancers. For example, Wang et al. found that has-miR-429 inhibits proliferation through the NF-κB pathway and suppresses cell migration-mediated epithelial-mesenchymal transition (EMT) in esophageal squamous cell carcinoma (26). It has also been confirmed that the overexpression of miRNA-429 inhibits proliferation and invasion in glioblastoma (27). Regarding reports on miRNA-29a-3p, Liao et al. stated that BLACAT1/miR-29a-3p/DVL3 promotes prostate cancer cell proliferation, migration, and invasion (28); moreover, miR-29a-3p is considered to be associated with gastric cancer (29). In addition, it has been proved that the NEAT1/miR-200b-3p/SMAD2 axis promotes melanoma progression (30), and miR-200b-3p regulates angiogenesis in hepatocellular carcinoma (31).
In addition, biglycan (BGN) is an important component of the ECM. Some studies have shown that solid tumors with a poor prognosis are often accompanied by upregulation of BGN (32-34). A great deal of research has shown that the ECM is a key regulator of cell and tissue function. Collagens are major components of the ECM. It has been reported that collagens impact the proliferation, invasion, initiation, metastasis, and therapy response in tumors (35). TGFBI, MMP7, and SERPINH1 have been found to be involved in tumor progression (36-38).
Of note, previous studies have confirmed the key roles of CCDC144NL-AS1, ZEB1-AS1, RRN3P2, MSC-AS1, and LINC01303 in cancer. For instance, He et al. (39) found that CCDC144NL-AS1 promoted the oncogenicity of osteosarcoma, Ma et al. (40) reported that downregulation of ZEB1-AS1 repressed cell proliferation, migration, and invasion in prostate cancer by mediating PI3K/AKT/mTOR signaling, Hu et al. (41) demonstrated that MSC-AS1 activated the Wnt/β-catenin signaling pathway to modulate cell proliferation and migration in renal clear cell carcinoma, and Cao et al. (42) found that the LINC01303/miR-101-3p/EZH2 axis promoted gastric cancer progression. Perhaps by modifying and adjusting these RNAs, it is possible to help in the treatment of tumors.
Inevitably, the current study had some limitations. First, our conclusions were mainly based on the ceRNA hypothesis, which requires future testing and verification. Second, the omission of significant potential information in the prediction of lncRNAs by combining the prognosis results with the hypothesis of ceRNA might have led to inaccurate results. Third, the analysis of the relationship between clinical characteristics and genes was absent.
Conclusions
In conclusion, the hub genes were associated with gastric adenocarcinoma, and a ceRNA network containing 24 mRNA-miRNA-lncRNAs was constructed based on the ceRNA hypothesis. Each component of the ceRNA network had a significant prognostic value in gastric adenocarcinoma and provides more evidence for future research on tumor markers and therapeutic targets in gastric adenocarcinoma.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1201/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1201/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Smyth EC, Nilsson M, Grabsch HI, et al. Gastric cancer. Lancet 2020;396:635-48. [Crossref] [PubMed]
- Zhang XY, Zhang PY. Gastric cancer: somatic genetics as a guide to therapy. J Med Genet 2017;54:305-12. [Crossref] [PubMed]
- Digklia A, Wagner AD. Advanced gastric cancer: Current treatment landscape and future perspectives. World J Gastroenterol 2016;22:2403-14. [Crossref] [PubMed]
- Shenouda SK, Alahari SK. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev 2009;28:369-78. [Crossref] [PubMed]
- Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016;29:452-63. [Crossref] [PubMed]
- Chan JJ, Tay Y. Noncoding RNA:RNA Regulatory Networks in Cancer. Int J Mol Sci 2018;19:1310. [Crossref] [PubMed]
- Qi X, Zhang DH, Wu N, et al. ceRNA in cancer: possible functions and clinical implications. J Med Genet 2015;52:710-8. [Crossref] [PubMed]
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353-8. [Crossref] [PubMed]
- Chen X, Leung SY, Yuen ST, et al. Variation in gene expression patterns in human gastric cancers. Mol Biol Cell 2003;14:3208-15. [Crossref] [PubMed]
- Cho JY, Lim JY, Cheong JH, et al. Gene expression signature-based prognostic risk score in gastric cancer. Clin Cancer Res 2011;17:1850-7. [Crossref] [PubMed]
- Cui J, Chen Y, Chou WC, et al. An integrated transcriptomic and computational analysis for biomarker identification in gastric cancer. Nucleic Acids Res 2011;39:1197-207. [Crossref] [PubMed]
- D'Errico M, de Rinaldis E, Blasi MF, et al. Genome-wide expression profile of sporadic gastric cancers with microsatellite instability. Eur J Cancer 2009;45:461-9. [Crossref] [PubMed]
- Wang Q, Wen YG, Li DP, et al. Upregulated INHBA expression is associated with poor survival in gastric cancer. Med Oncol 2012;29:77-83. [Crossref] [PubMed]
- Wei L, Sun J, Zhang N, et al. Noncoding RNAs in gastric cancer: implications for drug resistance. Mol Cancer 2020;19:62. [Crossref] [PubMed]
- Shin VY, Chu KM. MiRNA as potential biomarkers and therapeutic targets for gastric cancer. World J Gastroenterol 2014;20:10432-9. [Crossref] [PubMed]
- Yu C, Chen J, Ma J, et al. Identification of Key Genes and Signaling Pathways Associated with the Progression of Gastric Cancer. Pathol Oncol Res 2020;26:1903-19. [Crossref] [PubMed]
- Coussens LM, Werb Z. Inflammation and cancer. Nature 2002;420:860-7. [Crossref] [PubMed]
- Wang B, Zhang K, Meng S, et al. LncRNA-NONHSAT024778 promote the proliferation and invasion of chordoma cell by regulating miR-1290/Robo1 axis. Int J Biol Sci 2021;17:796-806. [Crossref] [PubMed]
- Liu Y, Xu G, Li L. LncRNA GATA3-AS1-miR-30b-5p-Tex10 axis modulates tumorigenesis in pancreatic cancer. Oncol Rep 2021;45:59. [Crossref] [PubMed]
- Zhou Z, Tan F, Pei Q, et al. lncRNA SNHG4 modulates colorectal cancer cell cycle and cell proliferation through regulating miR-590-3p/CDK1 axis. Aging (Albany NY) 2021;13:9838-58. [Crossref] [PubMed]
- Chen M, Wei X, Shi X, et al. LncRNA HIF1A-AS2 accelerates malignant phenotypes of renal carcinoma by modulating miR-30a-5p/SOX4 axis as a ceRNA. Cancer Biol Med 2021;18:587-603. [Crossref] [PubMed]
- Yang XZ, Cheng TT, He QJ, et al. LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/β-catenin pathway. Mol Cancer 2018;17:126. [Crossref] [PubMed]
- Zhang G, Li S, Lu J, et al. LncRNA MT1JP functions as a ceRNA in regulating FBXW7 through competitively binding to miR-92a-3p in gastric cancer. Mol Cancer 2018;17:87. [Crossref] [PubMed]
- Chen X, Chen Z, Yu S, et al. Long Noncoding RNA LINC01234 Functions as a Competing Endogenous RNA to Regulate CBFB Expression by Sponging miR-204-5p in Gastric Cancer. Clin Cancer Res 2018;24:2002-14. [Crossref] [PubMed]
- Wang Y, Yu XJ, Zhou W, et al. MicroRNA-429 inhibits the proliferation and migration of esophageal squamous cell carcinoma cells by targeting RAB23 through the NF-κB pathway. Eur Rev Med Pharmacol Sci 2020;24:1202-10. [PubMed]
- Peng G, Liao Y, Shen C. miRNA-429 Inhibits Astrocytoma Proliferation and Invasion by Targeting BMI1. Pathol Oncol Res 2017;23:369-76. [Crossref] [PubMed]
- Liao B, Chen S, Li Y, et al. LncRNA BLACAT1 Promotes Proliferation, Migration and Invasion of Prostate Cancer Cells via Regulating miR-29a-3p/DVL3 Axis. Technol Cancer Res Treat 2021;20:1533033820972342. [Crossref] [PubMed]
- Qu F, Zhu B, Hu YL, et al. LncRNA HOXA-AS3 promotes gastric cancer progression by regulating miR-29a-3p/LTβR and activating NF-κB signaling. Cancer Cell Int 2021;21:118. [Crossref] [PubMed]
- Zhou WJ, Wang HY, Zhang J, et al. NEAT1/miR-200b-3p/SMAD2 axis promotes progression of melanoma. Aging (Albany NY) 2020;12:22759-75. [Crossref] [PubMed]
- Moh-Moh-Aung A, Fujisawa M, Ito S, et al. Decreased miR-200b-3p in cancer cells leads to angiogenesis in HCC by enhancing endothelial ERG expression. Sci Rep 2020;10:10418. [Crossref] [PubMed]
- Ruan T, Lu S, Xu J, et al. lncRNA LINC00460 Functions as a Competing Endogenous RNA and Regulates Expression of BGN by Sponging miR-149-5p in Colorectal Cancer. Technol Cancer Res Treat 2021;20:1533033820964238. [Crossref] [PubMed]
- Huang HC, Cai BH, Suen CS, et al. BGN/TLR4/NF-B Mediates Epigenetic Silencing of Immunosuppressive Siglec Ligands in Colon Cancer Cells. Cells 2020;9:397. [Crossref] [PubMed]
- Chen D, Qin Y, Dai M, et al. BGN and COL11A1 Regulatory Network Analysis in Colorectal Cancer (CRC) Reveals That BGN Influences CRC Cell Biological Functions and Interacts with miR-6828-5p. Cancer Manag Res 2020;12:13051-69. [Crossref] [PubMed]
- Egeblad M, Rasch MG, Weaver VM. Dynamic interplay between the collagen scaffold and tumor evolution. Curr Opin Cell Biol 2010;22:697-706. [Crossref] [PubMed]
- Chiavarina B, Costanza B, Ronca R, et al. Metastatic colorectal cancer cells maintain the TGFβ program and use TGFBI to fuel angiogenesis. Theranostics 2021;11:1626-40. [Crossref] [PubMed]
- Liao HY, Da CM, Liao B, et al. Roles of matrix metalloproteinase-7 (MMP-7) in cancer. Clin Biochem 2021;92:9-18. [Crossref] [PubMed]
- Tian S, Peng P, Li J, et al. SERPINH1 regulates EMT and gastric cancer metastasis via the Wnt/β-catenin signaling pathway. Aging (Albany NY) 2020;12:3574-93. [Crossref] [PubMed]
- He J, Guan J, Liao S, et al. Long Noncoding RNA CCDC144NL-AS1 Promotes the Oncogenicity of Osteosarcoma by Acting as a Molecular Sponge for microRNA-490-3p and Thereby Increasing HMGA2 Expression. Onco Targets Ther 2021;14:1-13. [Crossref] [PubMed]
- Ma T, Chen H, Wang P, et al. Downregulation of lncRNA ZEB1-AS1 Represses Cell Proliferation, Migration, and Invasion Through Mediating PI3K/AKT/mTOR Signaling by miR-342-3p/CUL4B Axis in Prostate Cancer. Cancer Biother Radiopharm 2020;35:661-72. [Crossref] [PubMed]
- Hu Z, Li L, Cheng P, et al. lncRNA MSC-AS1 activates Wnt/β-catenin signaling pathway to modulate cell proliferation and migration in kidney renal clear cell carcinoma via miR-3924/WNT5A. J Cell Biochem 2020;121:4085-93. [Crossref] [PubMed]
- Cao C, Xu Y, Du K, et al. LINC01303 functions as a competing endogenous RNA to regulate EZH2 expression by sponging miR-101-3p in gastric cancer. J Cell Mol Med 2019;23:7342-8. [Crossref] [PubMed]




