
NDRG2 inhibition of glycolysis in liver tumor cells is regulated by SIRT1
Highlight box
Key findings
• We find that NDRG2 can inhibit glycolysis in liver tumor cells via SIRT1.
What is known and what is new?
• We know that NDRG2 is a tumor suppressor, but the function of NDRG2 in liver tumor glycolysis is completely unknown.
• This manuscript reported that SIRT1 is a key regulator involved in glycolysis in liver tumor cells.
What is the implication, and what should change now?
• This study findings enrich the understanding of the role of NDRG2 in tumor growth and of the mechanism by which NDRG2 regulates glycolysis.
Introduction
Liver tumor ranks among the top three malignant tumors due to their high heterogeneity, high mortality, strong invasiveness, difficult cure, and poor prognosis (1).
N-Myc downstream-regulated gene 2 (NDRG2) is a known tumor suppressor that is downregulated or even absent in various types of cancer (2,3). Overexpression of NDRG2 can inhibit the growth, and promote the apoptosis of cancer cells (4). Moreover, in liver tumors, researchers have demonstrated that overexpression of NDRG2 can inhibit cell migration and invasion by downregulating CD24 expression (5) and matrix metalloproteinases (MMPs) (6,7). All these reports prove the correlation between NDRG2 and the prognosis of liver tumors.
Glycolysis is a hallmark of cancer cells after metabolic reprogramming (8,9). Two studies reported the regulatory role of NDRG2 on glycolysis in colorectal cancer cells (10,11). Overexpression of NDRG2 in colorectal cancer cells reduced glucose consumption, lactate production, and increased oxygen consumption; knockdown of NDRG2 increased glucose consumption, lactate production increased, and decreased oxygen consumption, suggesting that NDRG2 has an inhibitory effect on glycolysis in tumor cells. Studies show that NDRG2 reduces the proliferation of colorectal cancer cells, possibly by downregulating glucose transport and metabolism-related enzymes such as glycolysis-related hexokinase 2 (HK2), pyruvate kinase M2 isoform (PKM2), lactate dehydrogenase A (LDHA) and GLUT1, and upregulating the expression of TNXIP (10,11). Shi et al demonstrated that NDRG2 inhibits glycolysis in clear-cell renal cell carcinoma in the same way (12).
Silent mating type information regulator 1 (SIRT1) belongs to the Sirtuin family and is an NAD+-dependent histone deacetylase (13-15). SIRT1-mediated deacetylation inhibits the functions of multiple tumor suppressors, including P53 (16), P73 (17), and HIC1 (18), suggesting that SIRT1 promotes tumorigenesis and progression. Emerging studies have shown that tumor progression affected by SIRT1 may be an important way of regulating glycolysis. SIRT1 stimulates the expression of glycolysis-related genes, such as GLUT1 and GAPDH, thus promoting glycolysis in tumors (19-21). Furthermore, SIRT1 can also interact with GAPDH and retains it in the cytosol, thus protecting the enzyme from nuclear translocation, and promoting glycolysis (22). Based on these reports, a possible correlation between NDRG2 and SIRT1 attracted our attention.
In the present study, we focused on the role of NDRG2 in regulating glycolysis and in regulating SIRT1 expression in liver tumor cells. Our data indicated a novel pathway of NDRG2 regulation of glycolysis in liver tumors. We present the following article in accordance with the MDAR reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-149/rc).
Methods
Patient information and tissue specimens
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of the Second Affiliated Hospital of Air Force Medical University (No. GKJ-Y-140) and informed consent was taken from all the patients. Fresh liver cell carcinoma specimens were collected from tumors resected from 143 patients at the Second Affiliated Hospital of Air Force Medical University (Xi’an, China) from 2004 to 2008. Liver tumor tissues and corresponding non-carcinoma tissues were confirmed by pathological review and staged according to the American Joint Cancer Committee/Union for International Cancer Control (AJCC/UICC) classification guidelines. The grading and histopathology subtyping of liver tumor specimens was based on the WHO criteria.
Immunohistochemistry
Immunohistochemical staining was performed to assess the protein expression of NDRG2 as described previously (23). Formalin-fixed tumor tissues were embedded in paraffin, and serial 4-mm sections were obtained using a Leica microtome. For staining, tumor sections were dewaxed in toluene, rehydrated in an alcohol gradient, permeabilized in citrate buffer (pH 6.0), quenched with 3% H2O2 for 5 min to eliminate endogenous peroxidase activity, washed in phosphate-buffered saline (PBS), incubated overnight with different antibodies and then with biotinylated goat anti-rat or anti-rabbit IgG antibody for 15 min. After washing, sections were incubated with streptavidin peroxidase, lightly counterstained with hematoxylin, and observed under a photomicroscope.
Cell lines and cell culture
HepG2 and SMMC-7721 and HEK-293T cells were purchased from Merck Millipore (Billerica, MA, USA) and cultured in Dulbecco’s modified Eagle’s medium (Invitrogen Life Technologies, USA) supplemented with 10% fetal bovine serum (FBS, Invitrogen Life Technologies) at 37 ℃ in an incubator (5% CO2, 21% O2 and 74% N2).
Lentivirus generation and infection
Recombinant lentiviral vectors were constructed with Invitrogen’s ViraPower™ Lentiviral System in our laboratory. The cDNAs of human NDRG2 were cloned and subcloned into the vector pLenti6. Short hairpin RNAs (shRNA) against human NDRG2 were designed using a small interfering RNA design program and then subcloned into the EcoR I/Age I sites of the pLKO-TRC vector. The shRNA sequences specific for NDRG2 were: shNDRG2-1 forward: 5'-CCGGGAGGACATGCAGGAAATCATTCTCGAGAATGATTTCCTGCATGTCCTCTTTTTG-3'; shNDRG2-1 reverse: 5'-AATTCAAAAAGAGGACATGCAGGAAATCATTCTCGAGAATGATTTCCTGCATGTCCTC-3'; shNDRG2-2 forward: 5'-CGGGATCCAAAAAAGCCACCTCAAGCGTCCGTCCTAGCAACAGCAAGCTTCCTGTTGCCAGGACAGACGCCTGAGGCGGCGGTGTTTCGTCCTTTCCACAA-3'; shNDRG2-2 reverse: 5'-CCCTCGAGCCCCAGTGGAA-3'. The sequences for the control nonsense shRNA were: control forward: 5'-CCGGAAGGTCTTGTCCTCATCAACACTCGAGTGTTGATGAGGACAAGACCTTTTTTTG-3'; control reverse: 5'-AATTCAAAAAAAGGTCTTGTCCTCATCAACACTCGAGTGTTGATGAGGACAAGACCTT-3'.
The HEK-293T cells were transfected with the pLenti6-Cherry/NDRG2, pLKO-Scramble/NDRG2-shRNA, PAX2, and PMD2G lentiviral vectors using Lipofectamine 2000 according to the manufacturer’s instructions (Invitrogen). After 48 h, the lentiviral supernatants were collected, filtered (0.45-µm size filter; Millipore, Billerica, MA, USA), and added to the HepG2/SMMC-7721 cells in the presence of 2 µg/mL Blasticidin (Sigma-Aldrich, USA) or 1 µg/mL Polybrene (Sigma-Aldrich, USA) for 6–8 h. Two rounds of infection were performed. After infection, the cells that survived this treatment were selected and collected after1 week had passed, and then analyzed for NDRG2 expression by Western blot.
Western blot analysis
The cells were harvested from 60-mm culture dishes and prepared by lysis in 200 mL RIPA buffer [0.05 M Tris-HCl (pH 7.4), 0.15 M NaCl, 0.25% deoxycholic acid, 1% Nonidet P-40, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 10 mg/mL aprotinin and 10 mg/mL leupeptin], supplemented with 100:1 (v/v) ratio of a protease inhibitor cocktail and phosphatase inhibitor cocktail. The protein concentrations in the lysates were measured using the bicinchoninic acid (BCA) protein assay. The proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes, which were saturated with Tris-buffered saline with 0.1% Tween 20 and 3% bovine serum albumin (TBST-BSA) and were then probed with the appropriate antibodies: NDRG2 (1:2,000, Cell Signaling Technology, USA), β-actin (1:2,000, Cell Signaling Technology, USA), SIRT1 (1:1,000, Cell Signaling Technology, USA), followed by incubation with species-matched secondary antibodies. The bands were detected using enhanced chemiluminescence (Pierce Rockford, USA) or the Odyssey Imaging System (LiCor Biosciences). The band intensities were quantified with Kodak Digital Science 1D 3.0 (Eastman Kodak, USAT).
Measurement of glucose uptake, lactate production, lactate dehydrogenase (LDH) activity and oxygen consumption rate
Cells were seeded on 6-well plates at a density of 2×105 cells per well and incubated at 37 ℃ for 24 h. The concentrations of glucose and lactate in the culture medium were measured by Glucose TestKit (Invitrogen) and Lactate Assay Kit (Jiancheng Bioengineering, China) respectively. Harvested cells were digested with 0.25% trypsin and washed with PBS. The cell suspension was homogenized on ice. LDH activity was measured by colorimetric assay using a specific test kit (Solarbio, China) according to the manufacturer’s instructions. The absorbance of LDH was measured at 450 nm and its activity in the control group was normalized to 1.0.
The cellular activity and oxygen consumption rate (OCR) were determined using the Seahorse XFe 96 Extracellular Flux Analyzer (Seahorse Bioscience, USA). Experiments were performed according to the manufacturer’s instructions. OCR was measured with the Seahorse XF Cell Mito Stress Test Kit (Seahorse Bioscience). Briefly, 2×105 cells were plated onto Seahorse plates, maintained overnight, and then washed with Seahorse buffer. Next, Seahorse buffers including oligomycin (Oligo), p-trifluoromethoxy carbonyl cyanide phenylhydrazone (FCCP), and rotenone + antimycin A (Rot + AA) were sequentially injected. The results were analyzed using software XF-96 wave (Seahorse Bioscience). All experiments were repeated at least 3 times.
Statistical analysis
The chi-square test or Fisher’s exact test and Student’s t-test were utilized to determine the significance of the differences between groups. The survival rates were analyzed using Kaplan-Meier analysis and log-rank test. The t-test method was used to compare the differences between groups. Statistical analysis was performed using SPSS software, Version 16.0 (Chicago, USA). Statistical significance was based on P<0.05.
Results
NDRG2 expression in liver tumor cells
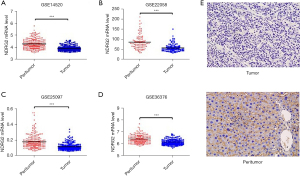
We first analyzed NDRG2 mRNA levels in patients using Gene Expression Omnibus (GEO), a database repository of gene expression profiles. As shown in Figure 1A-1D, the mRNA level of NDRG2 (P<0.05) was significantly decreased in liver tumors compared with peritumor from four sub-databases (GSE14520, GSE22058, GSE25097, GSE36376). The protein level of NDRG2 was assessed in 143 liver carcinoma tissues and corresponding non-carcinoma tissues. The results of immunohistochemical staining showed distinct staining of the cytoplasm of non-carcinoma tissues, but only faint or no staining in the liver tumor tissues (Figure 1E). Thus, we concluded that NDRG2 as a tumor suppressor was downregulated at both the mRNA and protein levels in liver tumors.
Correlation between NDRG2 expression and prognosis of liver tumor patients
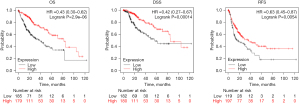
We then investigated the correlation between NDRG2 expression and survival status of liver tumor patients using Kaplan-Meier curves. Figure 2 shows that patients with higher NDRG2 expression had good overall survival (OS) (P<0.001), disease-specific survival (DSS) (P<0.001), and relapse-free survival (RFS) (P<0.01) compared with those with lower NDRG2 expression. In detail, the mean OS of patients with high and low NDRG2 protein expression levels were 81.9 and 38.3 months, respectively. The mean DSS of patients with high and low NDRG2 protein expression levels were 84.7 and 56.5 months, respectively. The mean RFS of patients with high and low NDRG2 protein expression levels were 37.2 and 15.17 months, respectively.
NDRG2 inhibits glycolysis in liver tumor cells
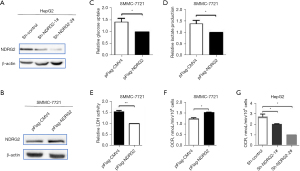
Glycolysis is a major feature of tumor cell metastasis (8,9). To prove whether NDRG2 can also inhibit glycolysis in liver tumor cells as in colorectal cancer cells (10), we constructed two liver tumor cell lines with stable knockdown and overexpression of NDRG2 by using an NDRG2-shRNA plasmid and NDRG2 eukaryotic expression plasmid, respectively (Figure 3A,3B). According to our data, overexpression of NDRG2 in the liver tumor cell line (SMMC-7721) inhibited glycolysis as suggested by decreased glucose uptake rate (Figure 3C), decreased lactate production (Figure 3D), decreased LDH activity (Figure 3E), and increased OCR (Figure 3F). Furthermore, NDRG2 knockdown by shRNA in liver tumor cells (HepG2) promoted glycolysis as indicated by decreased OCR (Figure 3G). Taken together, these data demonstrated that NDRG2 inhibited glycolysis in liver tumor cells.
Downregulation of SIRT1 by NDRG2
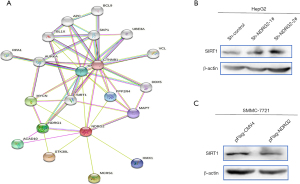
To further explore the role of NDRG2 in glucose metabolism, the protein interaction network of NDRG2 was achieved using the STRING database (https://string-db.org/) (Figure 4A). Impressively, SIRT1 was found in this network despite unknown or predicted interactions with NDRG2. Hence, we conceived a correlation between NDRG2 and SIRT1(24) that may contribute to glucose metabolism in liver tumor cells. Subsequent western blot analysis showed increased SIRT1 expression in NDRG2 knockdown cells but a decrease in NDRG2-overexpressed cells (Figure 4B,4C). Our experimental data suggested the expression of SIRT1 negatively correlated with that of NDRG2.
Discussion
NDRG2 is a known tumor suppressor involved in energy metabolism, especially glucose metabolism (25,26). In this study, the mRNA and protein expression levels of NDRG2 were analyzed in liver carcinoma tissues and non-carcinoma tissues, with the results reconfirming its downregulated expression in liver tumors (Figure 1).
Recently, it was reported that NDRG2 is involved in cellular glucose metabolism through insulin signal transduction based on NDRG2 being a substrate of kinase Akt and serum- and glucocorticoid-induced kinase 1 (SGK1) (27,28). According to the Warburg effect, cancer cells are more prone to glycolysis than oxidative phosphorylation for glucose metabolism (29,30). Glycolysis contributes to cancer progression (29), and glycolysis inhibition is emerging as a promising area of cancer therapy (31). To date, including in our present study, NDRG2 has been shown to significantly inhibit glycolysis of tumor cells in colorectal cancer (10,11), clear cell renal cell carcinoma (12), and liver tumor (Figure 3C-3F). In light of these reports, the suppression of glycolysis by NDRG2 is through the regulation of glycolysis-related genes. NDRG2 was first identified to decrease glucose uptake in breast cancer by promoting GLUT1 protein degradation without affecting GLUT1 transcription (32). Subsequently, the expressions of glycolysis-related hexokinase 2 (HK2), pyruvate kinase M2 isoform (PKM2), and lactate dehydrogenase A (LDHA) were proved to be significantly suppressed by NDRG2 in colorectal cancer cells and clear cell renal cell carcinoma cells (10,12). Moreover, NDRG2 could stimulate TXINP expression to reduce glucose uptake (11).
Interestingly, although NDRG2 is an N-My downstream-regulated gene, it is not repressed by transcription factor N-Myc but by C-Myc (10,33). C-Myc is a known viral oncogene in cancer energy metabolism (34,35) and mainly promotes the glycolysis of cancer cells through upregulating glycolysis gene expressions, such as LDH, HK2, GLUT1, and PKM2 (3). In addition, HIF-1 and P53, two other transcription factors, also play crucial roles in tumorigenesis by regulating the expression of glycolytic genes (36). HIF-1 promotes but P53 hinders these genes expression (37,38). Moreover, HIF-1 and P53 show negative and positive regulatory effects on NDRG2, respectively (39-42). Hence, we believe that the regulation by NDRG2 of glycolysis flux is accomplished by cooperating with C-Myc, HIF-1, and P53 to regulate the expression of glycolytic genes.
Sirtuins (e.g., SIRT1-7) play important roles in the Warburg effect and can regulate glycolytic genes through various effects (21). Sirtuins directly regulate the expression of glycolytic enzymes, alter the enzymatic activity of glycolytic genes via multiple post-translational modifications and affect the sub-location of the enzymes (19,21,43,44). For example, SIRT1promotes the expression of GLUT1, GAPDH, and LDHA to benefit glycolysis (19,43), interacts with GAPDH, and retains it in the cytosol, thus promoting glycolysis (22). NDRG2 and SIRT1 show opposing regulatory effects on glycolytic enzymes (Figure 4B,4C). The regulatory relationship between SIRT1 and glycolytic regulators is opposite to that between NDRG2 and glycolytic regulators. The expression of SIRT1 increases through C-Myc binding to the SIRT1 promoter, and then deacetylating C-Myc, and stimulating the transcriptional activity of C-Myc (21); SIRT1-mediated deacetylation suppresses the functions of P53 (45).
In this study, we found that SIRT1 regulates the inhibitory effects of NDRG2 on glycolysis in hepatocellular carcinoma cells, but further research on the relationship between SIRT1 and glycolysis is needed.
Conclusions
Overall, NDRG2 and SIRTI, as a pair of negative regulators, have opposing effects on tumor glycolysis, SIRT1 plays an important role in glycolysis regulation, is negatively regulated by NDRG2 in liver tumors.
Acknowledgments
Funding: This study was supported in part by the Natural Science Foundation of Shaanxi Province (No. 2023-YBSF-349).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-149/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-149/dss
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-149/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-149/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of the Second Affiliated Hospital of the Air Force Medical University (No. GKJ-Y-140) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen Z, Xie H, Hu M, et al. Recent progress in treatment of hepatocellular carcinoma. Am J Cancer Res 2020;10:2993-3036. [PubMed]
- Deng Y, Yao L, Chau L, et al. N-Myc downstream-regulated gene 2 (NDRG2) inhibits glioblastoma cell proliferation. Int J Cancer 2003;106:342-7. [Crossref] [PubMed]
- Hu W, Fan C, Jiang P, et al. Emerging role of N-myc downstream-regulated gene 2 (NDRG2) in cancer. Oncotarget 2016;7:209-23. [Crossref] [PubMed]
- Cao W, Zhang JL, Feng DY, et al. The effect of adenovirus-conjugated NDRG2 on p53-mediated apoptosis of hepatocarcinoma cells through attenuation of nucleotide excision repair capacity. Biomaterials 2014;35:993-1003. [Crossref] [PubMed]
- Zheng J, Li Y, Yang J, et al. NDRG2 inhibits hepatocellular carcinoma adhesion, migration and invasion by regulating CD24 expression. BMC Cancer 2011;11:251. [Crossref] [PubMed]
- Lee DC, Kang YK, Kim WH, et al. Functional and clinical evidence for NDRG2 as a candidate suppressor of liver cancer metastasis. Cancer Res 2008;68:4210-20. [Crossref] [PubMed]
- Guo Y, Ma J, Wu L, et al. Hyperthermia-induced NDRG2 upregulation inhibits the invasion of human hepatocellular carcinoma via suppressing ERK1/2 signaling pathway. PLoS One 2013;8:e61079. [Crossref] [PubMed]
- DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv 2016;2:e1600200. [Crossref] [PubMed]
- Luengo A, Gui DY, Vander Heiden MG. Targeting Metabolism for Cancer Therapy. Cell Chem Biol 2017;24:1161-80. [Crossref] [PubMed]
- Xu X, Li J, Sun X, et al. Tumor suppressor NDRG2 inhibits glycolysis and glutaminolysis in colorectal cancer cells by repressing c-Myc expression. Oncotarget 2015;6:26161-76. [Crossref] [PubMed]
- Hu J, Feng L, Ren M, et al. Colorectal Cancer Cell Differentiation Is Dependent on the Repression of Aerobic Glycolysis by NDRG2-TXNIP Axis. Dig Dis Sci 2022;67:3763-72. [Crossref] [PubMed]
- Shi W, Xu X, Yan F, et al. N-Myc downstream-regulated gene 2 restrains glycolysis and glutaminolysis in clear cell renal cell carcinoma. Oncol Lett 2017;14:6881-7. [Crossref] [PubMed]
- Kleszcz R, Paluszczak J, Baer-Dubowska W. Targeting aberrant cancer metabolism - The role of sirtuins. Pharmacol Rep 2015;67:1068-80. [Crossref] [PubMed]
- Giblin W, Skinner ME, Lombard DB. Sirtuins: guardians of mammalian healthspan. Trends Genet 2014;30:271-86. [Crossref] [PubMed]
- Mei Z, Zhang X, Yi J, et al. Sirtuins in metabolism, DNA repair and cancer. J Exp Clin Cancer Res 2016;35:182. [Crossref] [PubMed]
- Lamichane S, Baek SH, Kim YJ, et al. MHY2233 Attenuates Replicative Cellular Senescence in Human Endothelial Progenitor Cells via SIRT1 Signaling. Oxid Med Cell Longev 2019;2019:6492029. [Crossref] [PubMed]
- Dai JM, Wang ZY, Sun DC, et al. SIRT1 interacts with p73 and suppresses p73-dependent transcriptional activity. J Cell Physiol 2007;210:161-6. [Crossref] [PubMed]
- Hallows WC, Yu W, Denu JM. Regulation of glycolytic enzyme phosphoglycerate mutase-1 by Sirt1 protein-mediated deacetylation. J Biol Chem 2012;287:3850-8. [Crossref] [PubMed]
- Pinho AV, Mawson A, Gill A, et al. Sirtuin 1 stimulates the proliferation and the expression of glycolysis genes in pancreatic neoplastic lesions. Oncotarget 2016;7:74768-78. [Crossref] [PubMed]
- Chen J, Cao L, Li Z, et al. SIRT1 promotes GLUT1 expression and bladder cancer progression via regulation of glucose uptake. Hum Cell 2019;32:193-201. [Crossref] [PubMed]
- Zhu S, Dong Z, Ke X, et al. The roles of sirtuins family in cell metabolism during tumor development. Semin Cancer Biol 2019;57:59-71. [Crossref] [PubMed]
- Joo HY, Woo SR, Shen YN, et al. SIRT1 interacts with and protects glyceraldehyde-3-phosphate dehydrogenase (GAPDH) from nuclear translocation: implications for cell survival after irradiation. Biochem Biophys Res Commun 2012;424:681-6. [Crossref] [PubMed]
- Cai BL, Li Y, Shen LL, et al. Nuclear Multidrug Resistance-Related Protein 1 Is Highly Associated with Better Prognosis of Human Mucoepidermoid Carcinoma through the Suppression of Cell Proliferation, Migration and Invasion. PLoS One 2016;11:e0148223. [Crossref] [PubMed]
- Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev 2006;20:2913-21. [Crossref] [PubMed]
- Chen XL, Lei L, Hong LL, et al. Potential role of NDRG2 in reprogramming cancer metabolism and epithelial-to-mesenchymal transition. Histol Histopathol 2018;33:655-63. [PubMed]
- Kim G, Lim S, Kim KD. N-myc Downstream-Regulated Gene 2 (NDRG2) Function as a Positive Regulator of Apoptosis: A New Insight into NDRG2 as a Tumor Suppressor. Cells 2021;10:2649. [Crossref] [PubMed]
- Burchfield JG, Lennard AJ, Narasimhan S, et al. Akt mediates insulin-stimulated phosphorylation of Ndrg2: evidence for cross-talk with protein kinase C theta. J Biol Chem 2004;279:18623-32. [Crossref] [PubMed]
- Murray JT, Campbell DG, Morrice N, et al. Exploitation of KESTREL to identify NDRG family members as physiological substrates for SGK1 and GSK3. Biochem J 2004;384:477-88. [Crossref] [PubMed]
- Vaupel P, Schmidberger H, Mayer A. The Warburg effect: essential part of metabolic reprogramming and central contributor to cancer progression. Int J Radiat Biol 2019;95:912-9. [Crossref] [PubMed]
- Chen X, Xie C, Fan XX, et al. Novel direct AMPK activator suppresses non-small cell lung cancer through inhibition of lipid metabolism. Oncotarget 2017;8:96089-102. [Crossref] [PubMed]
- Sheng H, Tang W. Glycolysis Inhibitors for Anticancer Therapy: A Review of Recent Patents. Recent Pat Anticancer Drug Discov 2016;11:297-308. [Crossref] [PubMed]
- Ma J, Liu W, Guo H, et al. N-myc downstream-regulated gene 2 expression is associated with glucose transport and correlated with prognosis in breast carcinoma. Breast Cancer Res 2014;16:R27. [Crossref] [PubMed]
- Zhang J, Li F, Liu X, et al. The repression of human differentiation-related gene NDRG2 expression by Myc via Miz-1-dependent interaction with the NDRG2 core promoter. J Biol Chem 2006;281:39159-68. [Crossref] [PubMed]
- Dang CV, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res 2009;15:6479-83. [Crossref] [PubMed]
- Méndez-Lucas A, Li X, Hu J, et al. Glucose Catabolism in Liver Tumors Induced by c-MYC Can Be Sustained by Various PKM1/PKM2 Ratios and Pyruvate Kinase Activities. Cancer Res 2017;77:4355-64. [Crossref] [PubMed]
- Wu Z, Wu J, Zhao Q, et al. Emerging roles of aerobic glycolysis in breast cancer. Clin Transl Oncol 2020;22:631-46. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Li L, Liang Y, Kang L, et al. Transcriptional Regulation of the Warburg Effect in Cancer by SIX1. Cancer Cell 2018;33:368-85.e7. [Crossref] [PubMed]
- Wang L, Liu N, Yao L, et al. NDRG2 is a new HIF-1 target gene necessary for hypoxia-induced apoptosis in A549 cells. Cell Physiol Biochem 2008;21:239-50. [Crossref] [PubMed]
- Wang RX, Ou XW, Kang MF, et al. Association of HIF-1α and NDRG2 Expression with EMT in Gastric Cancer Tissues. Open Life Sci 2019;14:217-23. [Crossref] [PubMed]
- Liu N, Wang L, Li X, et al. N-Myc downstream-regulated gene 2 is involved in p53-mediated apoptosis. Nucleic Acids Res 2008;36:5335-49. [Crossref] [PubMed]
- Zhang K, Zhang Y, Zhang C, et al. Upregulation of P53 promotes nucleus pulposus cell apoptosis in intervertebral disc degeneration through upregulating NDRG2. Cell Biol Int 2021;45:1966-75. [Crossref] [PubMed]
- Mao B, Zhao G, Lv X, et al. Sirt1 deacetylates c-Myc and promotes c-Myc/Max association. Int J Biochem Cell Biol 2011;43:1573-81. [Crossref] [PubMed]
- Vettraino M, Manerba M, Govoni M, et al. Galloflavin suppresses lactate dehydrogenase activity and causes MYC downregulation in Burkitt lymphoma cells through NAD/NADH-dependent inhibition of sirtuin-1. Anticancer Drugs 2013;24:862-70. [Crossref] [PubMed]
- Lin Z, Fang D. The Roles of SIRT1 in Cancer. Genes Cancer 2013;4:97-104. [Crossref] [PubMed]
(English Language Editor: K. Brown)


