
RNA-binding protein ENO1 promotes the tumor progression of gastric cancer by binding to and regulating gastric cancer-related genes
Highlight box
Key findings
• First, ENO1 has a wide range of RNA-binding activities.
• Second, ENO1 may stabilize the expression of SOX9, VEGFA, GPRC5A, and MCL1 by binding to their mRNA, and promoting the development of gastric cancer.
• Third, ENO1 interacts with the long non-coding RNA NEAT1, LINC00511, CD44, and PKM, regulates their expression or alternative splicing, affects cell proliferation, migration, and apoptosis, and thus plays a role in GC.
What is known and what is new?
• With the exception of human epidermal growth factor receptor 2 (HER-2), there is no effective target for GC;
• In this study, we revealed a new mechanism of action whereby the RNA-binding protein ENO1 promotes glycolysis and tumor cell progression and identified a potential therapeutic target in GC.
What is the implication, and what should change now?
• Further research should be conducted to determine whether ENO1 affects the glycolysis of GC cells by activating the PI3K/AKT signaling pathway.
Introduction
With over 1 million new cases and an estimated 769,000 deaths in 2020, stomach cancer is the 5th most common cancer and 4th leading cause of cancer-related death worldwide (1). Most patients with gastric cancer (GC) are diagnosed in the late stages and have a poor prognosis and high mortality rate. At present, chemotherapy is still the main treatment for advanced GC, but neither changing the number of chemotherapy drugs, different combinations of chemotherapy drugs, nor the currently recommended immunotherapy can solve the high mortality rate of GC. So, more studies on the mechanisms underlying tumor progression need to be conducted due to the high heterogeneity of GC.
Glucose metabolism is critical in GC, and the enhancement of aerobic glycolysis has been shown to promote the occurrence, development, invasion, and metastasis of tumors (2). In our previous study, we showed that the overexpression of glycolytic pathway enolase 1 (ENO1), a key enzyme in glycolysis, promotes the proliferation of GC cells, and the survival rate of GC patients with high expression levels of ENO1 is lower than that of patients with low expression levels (3). Moreover, knocking down ENO1 in MKN-45 cells significantly inhibits the proliferation and clone formation of GC cells, promoting apoptosis and increasing their chemosensitivity (4). In-vivo studies on the effects of ENO1 on GC have shown that the deletion of ENO1 inhibits the tumorigenicity of GC cells in nude mice. Given that ENO1 is a key enzyme in glycolysis, we used the Seahorse XF96 extracellular flux analyzer to study the effects of ENO1 on the glycolytic ability of GC cells (4). By monitoring the extracellular acidification rate, we found that silencing ENO1 significantly reduced the glycolysis level of GC cells. Further, we found that the expression of ENO1 and pyruvate kinase M2 (PKM2), another key enzyme in the glycolytic pathway, was positively correlated in GC tissues (5). The correlation between the expression of ENO1 and PKM2 in the GC tissues was analyzed by the Oncomine database, and the messenger RNA (mRNA) expression correlation coefficient between them was as high as 0.886. Based on our previous findings and the literature, we showed that ENO1 is abnormally upregulated in GC and participates in the malignant development and glycolytic process of tumors. However, the role of ENO1 in this process remains unclear.
ENO1 is an RNA-binding protein (RBP) that specifically binds to RNA in HeLa cells and widely regulates the expression changes of RNA levels (6). A combination of ENO1 and long non-coding RNA (lncRNA) is involved in the occurrence of cancer. Zhang et al. (7) documented the mechanism by which ENO1 degrades mRNA as an RBP. They found that ENO1 protein binds to the mRNA of iron regulatory protein 1 (IRP1), and promotes the degradation of IRP1 mRNA by recruiting RNA degradation factor CNOT6. Combined with previous findings that ENO1 degrades RNA in prokaryotes, the research results reveal the conservation of ENO1 function among species. Notably, ENO1 binds to and degrades the mRNA of the IRP1 gene, thus regulating the metabolic homeostasis of ions in cells, affecting ferroptosis, and promoting the occurrence and development of liver cancer (7). Such findings have revealed a potential new target for the treatment of liver cancer-related diseases. However, the molecular mechanism of ENO1 as an RBP in the development of GC remains poorly understood.
In this study, we used RNA-immunoprecipitation sequencing (RIP-seq) to identify the pre-mRNA/mRNA sequences that bind to ENO1 in MKN-45 cells and also examined the molecular mechanism by which ENO1 interacts with other genes or pathways to promote the proliferation and metastasis of gastric tumor cells. We present the following article in accordance with the MDAR reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-151/rc).
Methods
Cell culture
The human GC cell line MKN45 was obtained from Procell (CL-0292, Wuhan, Hubei, China). ENO1 antibody was purchased from Abcam (Cambridge, UK).
Resuscitation of cells
A cryopreservation tube containing 1 mL of cell suspension was shaken quickly in a water bath at 37 ℃, and 4 mL of culture medium was added and mixed well. Centrifuged at 1,000 rpm for 4 minutes, the supernatant was discarded, 1–2 mL of culture medium was added and blown evenly. Next, all the cell suspensions were added to a culture flask and cultured overnight. The fluid was changed the next day and the cell density was checked. If the cell density reached 80–90%, a cell culture was carried out. The cells were cultured for 24 h in an incubator at 37 ℃ with 5% carbon dioxide (CO2) and 95% humidity for the subsequent experiments. The cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (HyClone, UT, USA) and cultured in an incubator at 37 ℃ with 5% CO2 and 95% humidity for 24 h.
Co-immunoprecipitation (IP)
The MKN45 cells were washed twice with cold 1× phosphate buffered saline (PBS). Formaldehyde was added to a final concentration of 1%, after which, it was gently mixed and incubated at room temperature for 10 min. To stop the crosslinking reaction, glycine was added at a final concentration of 0.125 M. The cells were harvested in cold PBS by scraping and were then transferred into a 1.5-mL microcentrifuge tube and centrifugated at 1,000 rpm for 5 min at 4 ℃. The collected cells were lysed in ice-cold wash buffer [1× PBS, 0.5% Sodium dodecyl sulfate (SDS), 0.5% NP-40, and 0.5% sodium deoxycholate] supplemented with 200 U/mL of RNase inhibitor (Takara) and protease inhibitor cocktail (Bimake) and incubated on ice for 30 min. The cell lysate was cleared by centrifugation at 10,000 rpm for 10 min at 4 ℃. RQI (Promega, 1 U/µL) was added to a final concentration of 0.1 U/µL and incubated in a water bath for 30 min at 37 ℃. Immediately afterwards, a stop solution was added to the lysates to quench the DNase. The mixture was then vibrated vigorously and centrifuged at 13,000 ×g at 4 ℃ for 20 min to remove the cell debris.
For the IP, the supernatant was incubated overnight at 4 ℃ with 10 µg of ENO1 antibody (Proteintech, 11204-1-AP) and control immunoglobulin G-antibody (CST, 2797s). The immunuprecipitates needed to incubate again for 2 h with protein A/G Dynabeads (Thermo Scientific Ltd.) at 4 ℃. After applying to magnets and removing the supernatants, beads were orderly washed by Tris-NaCl (pH 7.4) buffer (250 mM Tris, 750 mM NaCl, 10 mM SDS, 0.5% ethylene diamine tetraacetie acid (EDTA), 0.1% NP-40 and 0.5% deoxycholate), and PNK buffer (50 mM Tris, 20 mM EGTA and 0.5% NP-40), respectively and resuspend the beads in of Elution buffer (50 mM Tris 8.0, 10 mM EDTA and 1% SDS). A heat block was used for incubating the suspension to release the immunoprecipitated RBP with crosslinked vortex and RNA. Then, remove the magnetic beads and transfer into a clean microfuge tube. Proteinase K (Roche) was added into the supernatant (without immunoprecipitated) and immunoprecipitated RBP with crosslinked RNA, with final concentration of 1.2 mg/mL. Finally, the mixture was incubated for 120 min at 55 ℃ and the RNA was purified with Trizol reagent (Life technologies).
Library preparation
Complementary DNA libraries were prepared with the KAPA RNA Hyper Prep Kit (KAPA, KK8541) according to the manufacturer’s procedure. For the high-throughput sequencing, the libraries were prepared according to the manufacturer’s instructions, and the Illumina NovaSeq system was used for the 150 nucleotides (nt) paired-end sequencing.
Statistical analysis
After the reads were aligned onto the genome with HISAT2 (8), the unique comparison on the genome was obtained, and the comparison results of the polymerase chain reaction (PCR) duplicate were removed. Next, two software programs, Piranha and ABLIRC, were used to perform the peak labelling. Piranha has been described elsewhere (9). The ABLIRC strategy was used to identify the binding regions of GRCh38 on the genome as described previously (10). The process of peak labelling is described as follows. First, the whole genome was scanned with 5 bp as a window and 5 bp as a step from the beginning of each chromosome. A peak was identified if the depth of the first window was 2.5 times that of 8 consecutive windows on the genome or had a medium depth >50. When the 8 consecutive windows were <4% of the maximum depth of the peak, the peak ended. Additionally, the reads on each gene were randomly distributed to each gene 500 times, and the frequency of the depth of each peak was calculated to conduct a significance analysis of the identified peak and identify the significant peaks (i.e., P values <0.05) or those with a maximum depth of a certain degree (≥10). Next, with the input samples as the control, an abundance difference analysis was conducted on the locations of these peaks, and a peak with an IP abundance >4 times (adjustable parameter) that of the input abundance was identified as the final combination peak. The IP target genes were ultimately determined by the peaks, and the HOMER software was used to label the binding motifs of the IP protein (11).
Functional enrichment analysis
The Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were identified using the KOBAS 2.0 server (12). The hypergeometric test and Benjamini-Hochberg FDR controlling procedure were used to define the enrichment of each term.
Results
ENO1 gene has a wide range of RNA-binding activities, and its binding is significantly enriched in the 5' untranslated region (UTR), 3'UTR, coding sequence (CDS), and intron regions
An improved RIP-seq (iRIP-seq) analysis was conducted on the antibody against ENO1 in the human GC cell line (MKN-45), and high-throughput sequencing was conducted. After IP was repeated twice, protein signal of ENO1 in IP sample is detected, which proves that IP is successful; ENO1 protein can still be detected in the supernatant, which indicates that the ENO1 protein in the lysate has not been completely precipitated, and the IP was successful (Figure 1A-1D).
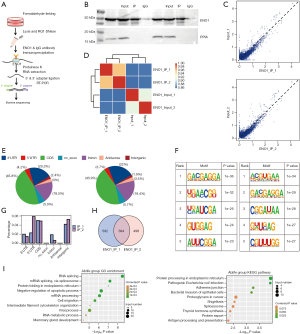
For the input sample, the IP sample reads were significantly enriched in the 5'UTR, 3'UTR, CDS, and intron regions, which showed that ENO1 has a wide range of RNA-binding activities (Figure 1E-1G). The binding motif of ENO1 obtained by the two experiments was consistent, and ENO1 was shown to bind to the GACGAGGA motif and CCAAG on the RNA (Figure 1F). The overlap of the ENO1 binding peaks obtained by the repeated experiments is shown in the following figure (Figure 1H). The peak-related genes were repeated in 2 experiments using the ABLIRC algorithm (software programs), clustered (Figure 1C), and underwent a GO function analysis (Figure 1I). Interestingly, we found that the consistency of the 2 experiments was very high, and both were significantly enriched in mRNA splicing and mRNA processing by splicing. The significantly enriched GO terms included RNA splicing, the viral process, the response to endoplasmic reticulum stress, the negative regulation of transcription by RNA polymerase II, the endoplasmic reticulum, protein folding, the negative regulation of apoptosis, cell migration, neutrophil degranulation, and other biological processes (Figure 1).
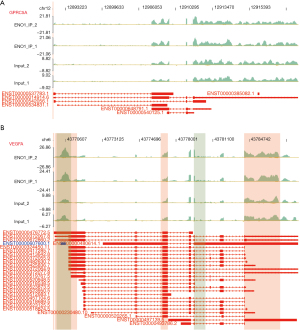
ENO1 can combine with lncRNA NEAT1 (Figure 2A), PKM (Figure 2B), CD44 (Figure 2C), and LINC00511 (Figure 3)
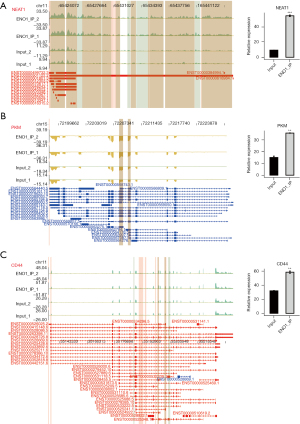
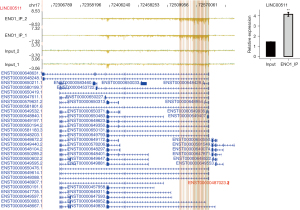
Among the top 30 ENO1-binding genes, NEAT1, LINC00511, CD44, and PKM appeared repeatedly in the IP-1 and IP-2 samples. ENO1 binds to many splicing factors, which suggests that it may indirectly regulate the expression of other genes by binding to the splicing factors. Notably, ENO1 can bind to its own mRNA. The above-mentioned genes are all related to GC.
The lncRNA NEAT1 plays an important role in the occurrence and progression of GC and may be used as a potential biomarker for treatment strategies and prognosis prediction. LINC00511 promotes the growth of GC cells by regulating competing endogenous RNA (ceRNA) and is also a promising therapeutic target for GC. CD44 is an important gene involved in the development of GC. PKM encodes a protein involved in glycolysis. The encoded protein is a pyruvate kinase (PK) that catalyzes phosphate groups. From phosphoenolpyruvic acid to adenosine diphosphate (ADP), adenosine triphosphate (ATP) and pyruvic acid are produced, and PKM was present in the data of the two IP repeats of ENO1 (Figures 2,3).
G protein-coupled receptor class C group 5 member A (GPRC5A), vascular endothelial growth factor A (VEGFA), myeloid cell leukemia-1 (MCL1), and SRY-box transcription factor 9 (SOX9) genes were differentially expressed genes (DEGs) that bind to ENO1
In the early stage, we knocked down ENO1 in the GC cell line MKN-45 and established a control group with 3 replicates per group (4). Using chip technology, we detected the transcriptome changes and screened 680 DEGs, of which 409 were downregulated, and 271 were upregulated [fold change (FC) >2 or FC <2, P<0.01]. The chip and iRIP-seq data were integrated and analyzed, and the intersection of the ENO1-binding genes and DEGs yielded 28 genes. Based on the expression and binding capacity of these 28 genes, 4 genes were ultimately selected by searching the literature on GC or cancer. It is speculated that ENO1 may bind to MCL1, SOX9, vascular GPRC5A, and VEGFA, and may play a role in stabilizing their expression. When ENO1 was silenced, its stabilizing effect on the transcription of these genes disappeared, promoting their degradation and resulting in decreased expression, and thus inhibiting the occurrence and development of GC (Figures 4,5).
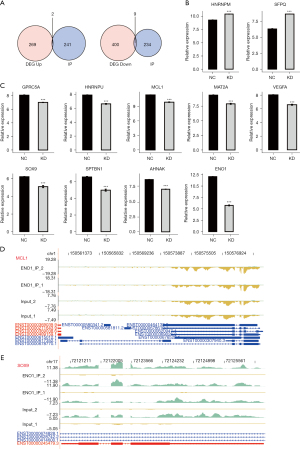
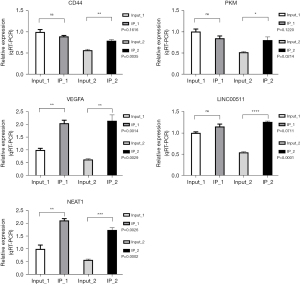
NEAT1, LINC00511, VEGFA, CD44 and PKM genes were selected for the RIP-PCR verification experiment
In IP-1, the VEGFA and NEAT1 genes were enriched and combined as expected, and there was a significant difference in the t-test results. The LINC00511 gene tended to be enriched. The CD44 and PKM genes are contrary to expectations, but there was no enrichment combination. In IP-2, the CD44, VEGFA, LINC00511, and NEAT1 genes were enriched and combined as expected, and there was a significant difference in the t-test results. Contrary to expectations, the PKM gene had no enrichment binding (Figure 6).
Discussion
Given the very high incidence and mortality of GC in China, in-depth analyses of the mechanism of glycolysis in GC and explorations of the corresponding targets and clinical applications are essential if the therapeutic effect and survival rate are to be improved. Previous studies have primarily focused on the occurrence, development, and transformation mechanism of GC. It has been established that changes in energy metabolism represent one of the main characteristics of tumors. In GC, the enhancement of aerobic glycolysis promotes the occurrence, development, invasion and metastasis, and is also related to poor patient prognosis.
RBPs are essential binding partners of intracellular RNA. They dynamically bind to RNA and can form a variety of complexes, including ribonucleoprotein particles. RBPs play an important role in regulating gene expression after transcription and regulate the function of cells by interacting with RNAs. RBPs are reportedly involved in various aspects of RNA metabolism, including RNA splicing, polyadenylation, sequence editing, RNA transport, RNA stability and degradation maintenance, intracellular localization, and translation control (13).
Enolase is a key enzyme in glycolysis, which catalyzes the dehydration of 2-phosphoglyceric acid and the formation of phosphoenolpyruvate (PEP) acid. ENO1 is known to accelerate cancer progression as a glycolytic enzyme; however, recent findings suggest that it may function as an RBP. RNA regulates glycolysis and embryonic stem cell differentiation via ENO1 (14). ENO1 is a member of the RNA degradosome in prokaryotes, it can bind to lncRNA and participate in the occurrence and progress of tumor (15). Moreover, Huang et al. (16) found that GC cell line MGC-803 knocked down the expression profile of ENO1 gene, as a result, there were 448 DEG, of which 183 (40.85%) were down-regulated. Deng et al. (17) found that CCDC65 as a new potential tumor suppressor induced by metformin inhibits activation of AKT1 via ubiquitination of ENO1 in GC. ENO1 widely regulates the expression of RNA in GC cells (18).
In this study, we conducted the iRIP-seq of ENO1 in MKN-45 cells and found that ENO1 binds to many coding protein genes and to the 3'UTR region, 5'UTR region, CDS region, and intron region of the genes, which shows the specific binding characteristics of this protein and indicates its function in regulating the alternative splicing of mRNA by binding to the GACGAGGA motif and CCAAG motif on RNA. The repeated peak-related genes in the 2 experiments were analyzed by a GO function analysis, and the biological processes related to RNA splicing and cell migration were significantly enriched (Figure 1). We found that ENO1 was combined with NEAT1, LINC00511, CD44, and PKM (Figures 2,3), which suggest that it might indirectly regulate the expression of these genes by binding to the splicing factors.
NEAT1 and LINC00511 are important lncRNAs involved in the development of GC. LncRNAs are a group of transcripts that can change the chromatin structure and regulate gene expression at the level of transcription and post-transcription, and the dysregulated expression of lncRNAs can lead to phenotype changes in GC cells (19). In 2007, Hutchinson et al. identified a long-chain non-coding RNA necessary for maintaining the completion of paraspeckle and named it NEAT1 (20). LncRNA NEAT1 is overexpressed in GC tissues and cell lines and is related to the clinical stage, histological type, lymph node metastasis, and distant metastasis. The survival rate of patients with high levels of lncRNA NEAT1 is lower than that of patients with low levels of lncRNA NEAT1. Alternatively, the knockdown of lncRNA NEAT1 has been shown to significantly inhibit the migration and invasion of GC cells in vitro and regulate the expression of epithelial mesenchymal transition (EMT)-related proteins. Thus, it has been established that lncRNA NEAT1 plays an important role in the occurrence and development of GC, and it may be used as a potential biomarker for treatment strategies and prognosis prediction (21). LINC00511 also promotes the growth of GC cells by regulating ceRNA and is also a promising therapeutic target for GC. In this study, we found that LINC00511 was highly expressed in each stage GC tissues and cell lines, which is consistent with previous research (22). Notably, the knockdown of LINC00511 has been shown to inhibit cell proliferation and increase the apoptosis rate in GC (23).
CD44 is a transmembrane glycoprotein and a surface receptor of hyaluronic acid. CD44 splice variants play a role in carcinogenesis, differentiation, and lymph node metastasis, and predict the prognosis of various cancers, including GC. Current evidence suggests that gastric stem cells express the splice variant CD44v9. Notably, CD44 and its splice variants are positively correlated with the occurrence and development of GC and may play an important role in diagnosis, treatment, and prognosis (24).
PKM is an important rate-limiting enzyme in the glycolytic pathway, which catalyzes the dephosphorylation of PEP to pyruvic acid. The isoenzymes PKM1 and PKM2 are encoded by the same precursor mRNA during transcription. In this experiment, PKM was present in the data of the 2 IP repeats of ENO1, which was consistent with our results. Thus, we conclude that ENO1 binds directly to PKM2, and speculate that ENO1 might directly bind to the pre-mRNA of PKM, and thus regulate the alternative splicing of PKM, leading to increased PKM2 expression and promoting glycolysis and the development of GC.
We also integrated our previous chip transcriptome data on the knockdown ENO1 in the GC cell line MKN-45 and conducted formaldehyde-crosslinking RIP-seq (fRIP-seq) of ENO1 in the MKN-45 cell line. We intersected the binding peak-related genes in the 2 repeated experiments of fRIP-seq and the DEGs obtained from the chip data of the ENO1 knockdown. Finally, we found that GPRC5A, VEGFA, MCL1, and SOX9 were DEGs that bind to ENO1. Thus, we speculate that ENO1 binds to SOX9, VEGFA, GPRC5A, and MCL1 and plays a role in stabilizing their expression. When ENO1 is silenced, its stabilizing effect on the transcription of these genes disappeared, and their degradation was accelerated, resulting in the occurrence and development of GC.
There are some limitations in this study. Our team found that ENO1 and PKM2 are directly related, and PKM also appeared in the data of two IP repeats of ENO1 in immunoprecipitation. However, this study has not yet reached a clear conclusion on how ENO1 differentially regulates alternative splicing of PKM2 and the subsequent differences in biological effects.
Conclusions
In summary, ENO1 has a wide range of RNA-binding activities, and its binding is significantly enriched in the 5'UTR, 3'UTR, CDS, and intron regions. The GO and KEGG enrichment analyses showed that ENO1 may regulate mRNA alternative splicing, and it binds to the RNA molecules that are mainly involved in mRNA splicing, mRNA processing, RNA splicing, virus processing, and responding to endoplasmic reticulum stress. The negative regulation of transcription by RNA polymerase II, the protein folding of endoplasmic reticulum, the negative regulation of apoptosis, cell migration, neutrophil degranulation and other biological processes. Additionally, ENO1 may further stabilize the expression of SOX9, VEGFA, GPRC5A, and MCL1 by binding to their mRNA, thus promoting the occurrence and development of GC. Third, ENO1 interacts with NEAT1, LINC00511, CD44, and PKM, regulates their expression or alternative splicing, affects cell proliferation, migration, and apoptosis, and thus plays a role in GC. Our findings have extended understandings of the mechanism of action of ENO1 as a clinical therapeutic target.
Acknowledgments
Funding: This work was supported by: (I) Project name: Mechanism and targeted therapy of ENO1 promoting aerobic glycolysis of gastric cancer cells by regulating PI3K/AKT/mTOR/PKM2 pathway (Grant No. 20YF3FA029; Category: Gansu Provincial Key Research and Development Project-Social Development Category; Source: Gansu Provincial Science and Technology Department). (II) Project name: Mechanism of activating PKM2 through VEGF ENO1/PI3K/AKT pathway to promote aerobic glycolysis of gastric cancer cells (Grant No. ZX-62000002-2021-139; Category: The Hospital Fund of Lanzhou University First Hospital; Source: Lanzhou University First Hospital). (III) The mechanism of ENO1 regulating aerobic glycolysis through PI3K/AKT/c-Fos pathway to promote the proliferation and metastasis of gastric cancer (Grant No. 20JR10RA672; Category: Gansu Science and Technology Project; Source: Gansu Provincial Science and Technology Department). (IV) Project name: Effect of ENO1 on the proliferation and metastasis of gastric cancer and its molecular mechanism (Grant No. 2020-25; Category: Young Scientific and Technological Talents Promotion Project in 2020 from Gansu Association for Science and Technology; Source: Gansu Province Science and Technology Association).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-151/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-151/dss
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-151/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-151/coif). All authors report that this work was supported by: (I) Project name: Mechanism and targeted therapy of ENO1 promoting aerobic glycolysis of gastric cancer cells by regulating PI3K/AKT/mTOR/PKM2 pathway (Grant No. 20YF3FA029; Category: Gansu Provincial Key Research and Development Project-Social Development Category; Source: Gansu Provincial Science and Technology Department). (II) Project name: Mechanism of activating PKM2 through VEGF ENO1/PI3K/AKT pathway to promote aerobic glycolysis of gastric cancer cells (Grant No. ZX-62000002-2021-139; Category: The Hospital Fund of Lanzhou University First Hospital; Source: Lanzhou University First Hospital). (III) The mechanism of ENO1 regulating aerobic glycolysis through PI3K/AKT/c-Fos pathway to promote the proliferation and metastasis of gastric cancer (Grant No. 20JR10RA672; Category: Gansu Science and Technology Project; Source: Gansu Provincial Science and Technology Department). (IV) Project name: Effect of ENO1 on the proliferation and metastasis of gastric cancer and its molecular mechanism (Grant No. 2020-25; Category: Young Scientific and Technological Talents Promotion Project in 2020 from Gansu Association for Science and Technology; Source: Gansu Province Science and Technology Association). The authors have no other conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Liu X, Wang X, Zhang J, et al. Warburg effect revisited: an epigenetic link between glycolysis and gastric carcinogenesis. Oncogene 2010;29:442-50. [Crossref] [PubMed]
- Zhou X, Yao K, Zhang L, et al. Identification of Differentiation-Related Proteins in Gastric Adenocarcinoma Tissues by Proteomics. Technol Cancer Res Treat 2016;15:697-706. [Crossref] [PubMed]
- Qiao H, Wang YF, Yuan WZ, et al. Silencing of ENO1 by shRNA Inhibits the Proliferation of Gastric Cancer Cells. Technol Cancer Res Treat 2018;17:1533033818784411. [Crossref] [PubMed]
- Ni TG, Gao C, Zhou X, et al. Expression of enolase-α and tumor M2 pyrurate kinase in gastric cancer and their clinical significances. Chinese Journal of Cancer Biotherapy 2011;18:524-7.
- Huppertz I, Attig J, D'Ambrogio A, et al. iCLIP: protein-RNA interactions at nucleotide resolution. Methods 2014;65:274-87. [Crossref] [PubMed]
- Zhang T, Sun L, Hao Y, et al. ENO1 suppresses cancer cell ferroptosis by degrading the mRNA of iron regulatory protein 1. Nat Cancer 2022;3:75-89. [Crossref] [PubMed]
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015;12:357-60. [Crossref] [PubMed]
- Uren PJ, Bahrami-Samani E, Burns SC, et al. Site identification in high-throughput RNA-protein interaction data. Bioinformatics 2012;28:3013-20. [Crossref] [PubMed]
- Xia H, Chen D, Wu Q, et al. CELF1 preferentially binds to exon-intron boundary and regulates alternative splicing in HeLa cells. Biochim Biophys Acta Gene Regul Mech 2017;1860:911-21. [Crossref] [PubMed]
- Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010;38:576-89. [Crossref] [PubMed]
- Xie C, Mao X, Huang J, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 2011;39:W316-22. [Crossref] [PubMed]
- Castello A, Fischer B, Hentze MW, et al. RNA-binding proteins in Mendelian disease. Trends Genet 2013;29:318-27. [Crossref] [PubMed]
- Huppertz I, Perez-Perri JI, Mantas P, et al. Riboregulation of Enolase 1 activity controls glycolysis and embryonic stem cell differentiation. Mol Cell 2022;82:2666-2680.e11. [Crossref] [PubMed]
- Hua Q, Wang D, Zhao L, et al. AL355338 acts as an oncogenic lncRNA by interacting with protein ENO1 to regulate EGFR/AKT pathway in NSCLC. Cancer Cell Int 2021;21:525. [Crossref] [PubMed]
- Huang Z, Lin B, Pan H, et al. Gene expression profile analysis of ENO1 knockdown in gastric cancer cell line MGC-803. Oncol Lett 2019;17:3881-9. [Crossref] [PubMed]
- Deng T, Shen P, Li A, et al. CCDC65 as a new potential tumor suppressor induced by metformin inhibits activation of AKT1 via ubiquitination of ENO1 in gastric cancer. Theranostics 2021;11:8112-28. [Crossref] [PubMed]
- Castello A, Hentze MW, Preiss T. Metabolic Enzymes Enjoying New Partnerships as RNA-Binding Proteins. Trends Endocrinol Metab 2015;26:746-57. [Crossref] [PubMed]
- Wen Y, Song XB, Ying BW. Research Progresses of miRNA and lncRNA in Gastric Cancer. Journal of Cancer Control and Treatment 2018;31:438-43.
- Hutchinson JN, Ensminger AW, Clemson CM, et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 2007;8:39. [Crossref] [PubMed]
- Fu JW, Kong Y, Sun X. Long noncoding RNA NEAT1 is an unfavorable prognostic factor and regulates migration and invasion in gastric cancer. J Cancer Res Clin Oncol 2016;142:1571-9. [Crossref] [PubMed]
- Sun CB, Wang HY, Han XQ, et al. LINC00511 promotes gastric cancer cell growth by acting as a ceRNA. World J Gastrointest Oncol 2020;12:394-404. [Crossref] [PubMed]
- Chen Z, Wu H, Zhang Z, et al. LINC00511 accelerated the process of gastric cancer by targeting miR-625-5p/NFIX axis. Cancer Cell Int 2019;19:351. [Crossref] [PubMed]
- Jang BI, Li Y, Graham DY, et al. The Role of CD44 in the Pathogenesis, Diagnosis, and Therapy of Gastric Cancer. Gut Liver 2011;5:397-405. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


