
Bromodomain-containing protein 9 activates proliferation and epithelial-mesenchymal transition of colorectal cancer via the estrogen pathway in vivo and in vitro
Highlight box
Key findings
• BRD9 may be an independent prognostic factor for CRC. The BRD9/estrogen pathway may contribute to the proliferation and EMT of CRC cells, suggesting a promising novel molecular target in the therapeutic strategy against CRC.
What is known and what is new?
• BRD9 has been reported in several cancers, such as prostate cancer, renal cell carcinoma, and synovial sarcoma.
• This investigation demonstrated that BRD9 might act via the estrogen pathway to affect the proliferation and EMT of CRC cells.
What is the implication, and what should change now?
• BRD9 might be a novel promising molecular target in the therapeutic management of CRC.
Introduction
Globally, colorectal cancer (CRC) is the third most commonly diagnosed cancer and is associated with the second largest number of cancer-related deaths (1). Indeed, its mortality is higher in women, especially in postmenopausal females, possibly due to the loss of estrogen in such women (2). However, the mechanisms through which estrogen and other biomarker influence the risk of advanced CRC in postmenopausal women is uncertain. Additionally, the TNM (tumor, node, metastasis) classification is limited because patients with the same pathological stage may have different prognosis (3). Therefore, continued interest in biomarkers as prognostic predictors allows more accurate patient stratification for curative resection of CRC.
Bromodomains are structural modules maintained in chromatin-bound proteins and acetyltransferase histones and are the only protein domains known to recognize acetyl-lysin residues on proteins (4). Bromodomain-containing proteins (BRD), which participate in gene fusions that generate diversity, are frequently dysregulated in cancer (5). However, many BRD subunits still require further investigation. Bromodomain-containing protein 9 (BRD9), a member of the bromodomain and extra terminal domain protein family, is a potential target for cancer, inflammatory diseases, and metabolic disorders (6). Wang et al. suggested that BRD9 may be involved in SWI/SNF transcription, DNA repair, and uncontrolled cellular differentiation (7). BRD9 knockdown or its antagonist suppressed the growth of clear cell renal cell carcinoma in vitro (8). BRD9 is also reported to have effects on several chemotherapies against leukemia (9). BRD9 and androgen receptor (AR) are co-dependent for binding select genes, which has been reported in other SWI/SNF-transcription factor partnerships (10). Additionally, BRD9 is believed to be a potential diagnostic and prognostic biomarker in prostate cancer (11). Moreover, BRD9-containing SWI/SNF subcomplex is required to survive SMARCB1-mutant rhabdoid tumors (7). In conclusion, the studies talked about above suggested the potential role of BRD9 in cancer progression, however, the literature has seldom studied the function and mechanism of BRD9 in colorectal cancer.
Therefore, this current study investigated the role of BRD9 in CRC.
Epithelial-to-mesenchymal transition (EMT), an essential biological mechanism of normal development, is characterized by loss of epithelial phenotype and conversion to mesenchymal phenotype, resulting in altered phenotype and function in cells. In patients with CRC, those with a high level of EMT have a greater tendency to metastasize to distant organs, such as the liver, compare patients with a lower level of EMT (12).
The estrogen signaling pathway is involved in the regulation of EMT in cancer cells (13). EMT is regulated by the target genes of the ERK, which are significantly important for the estrogen pathway (14). Yet, it remains unclear whether BRD9 is involved in the oncogenic process through the EMT/estrogen pathway.
The literatures have been reported that BRD9 might be an important biomarker in colon cancer (15,16), while their studies little investigate the clinical significance of BRD9 in colorectal cancer. Our study intended to explore the clinical significance of BRD9 in colorectal cancer. In addition, the relationship between BRD9 expression in CRC tissues and cell lines was examined. Moreover, we investigated the effects of BRD9 on the proliferation and EMT of CRC cells and discussed the potential molecular mechanisms. Finally, the influence of BRD9 knockdown on tumorigenesis was investigated in vivo. We present the following article in accordance with the MDAR and ARRIVE reporting checklists (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-271/rc).
Methods
Sample collection
A total of 31 fresh tissues of CRC and para-tumor from patients undergoing dissection surgery in the General Department of the Chinese PLA General Hospital between September 2022 and January 2022 were collected. Immunohistochemistry (IHC) staining, Western blot (WB), and quantitative real-time polymerase chain reaction (qRT-PCR) were used to investigate the expression of BRD9. A total of 524 CRC paraffin-embedded samples were gathered from the General Department between February 2010 and December 2015 and analyzed. The following patient characteristics were analyzed: age, sex, microsatellite instability (MSI), carcinoembryonic antigen (CEA), location of tumor, differentiation, tumor size, T stage, N stage, and TNM classification. Surgical procedures, including the extent of both colorectal resection and lymph node dissection, were based on the 2010 Japanese Society for Cancer of the Colon and Rectum (JSCCR) guidelines for the treatment of CRC. All 524 patients were followed for up to 60 months. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the ethical review committee of the Chinese PLA General Hospital (Approval No. S2019-228-02). Written informed consent was collected from all patients. Guanglong Dong was aware of the group allocation at the different stages of the experiment (during the allocation, the conduct of the experiment, the outcome assessment, and the data analysis).
Study outcomes
The primary outcome variable for this study was the recurrence of CRC, categorized as local or distant recurrence. Secondary outcomes included the expression of BRD9, location of tumor, differentiation of the tumor, tumor size, T stage, N stage, and TNM classification. For this analysis, we included the first recurrence that occurred between 90 days and 5 years from the definitive operation.
Cell lines and qRT-PCR
CRC cell lines HT-29, MC-38, SW480, LOVO, and HCT-116, and normal human colorectal mucosal epithelial cell line FHC were purchased from the Chinese Academy of Sciences and cultured in Dulbecco’s Modified Eagle Medium (DMEM; Shanghai, China). Total RNA was extracted from CRC cell lines and tissues using the TRIzol reagent and total RNA quality was determined using the Nanodrop 2,000 spectrophotometer (TermoFisher, MA, USA). The SYBR™ qPCR SuperMix (Novoprotein, Shanghai, China) was used to synthesis the cDNA and perform qRT-PCR. The expression levels of mRNAs were normalized to the expression of glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The primers used were as follows: BRD9 (forward) 5'-ATGTTCCATGAAGCCTCCAG-3' and BRD9 (reverse) 5'-AGCTCCTTCTTCACCTTCCC-3'; GAPDH (forward) 5'-GCGAGATCGCACTCATCATCT-3' and GAPDH (reverse) 5'-TCAGTGGTGGACCTGACC-3'. The qRT-PCR results were analyzed using the 2−∆∆Ct method, and each sample was repeated three times.
Lentivirus-transduced stable cell lines
A lentiviral plasmid with short hairpin RNA (shRNA) was designed to target human BRD9 and was purchased from JinTuoSi Co. Ltd. (Wuhan, China). A non-silent (vector) shRNA was used as the negative control. A lentiviral plasmid with overexpression RNA (oeBRD9) was designed to target human BRD9 and was purchased from JinTuoSi Co. Ltd. (Wuhan, China). A non-silent (vector) oeBRD9 was used as the negative control. Briefly, stable clones were generated by transfecting SW480 cells in a 6-well plate with 2 µg of each shRNA plasmid. At least 7 days after transfection, the cells were cultivated in a medium containing 2.5 µg/mL puromycin. The transfection efficiency was verified by qRT-PCR and Western blot experiments.
Protein extraction and Western blot
The cells or tissues were resuspended in RIPA buffer (Beyotime, Shanghai, China) and kept on ice for 30 minutes, centrifuged at 15,000 r/min for 15 minutes at 4 ℃, and the supernatant was extracted. The total protein was detected using the BCA protein assay kit (Solarbio, Beijing, China) and samples were boiled at 95 ℃ for 5 minutes. The proteins were separated via sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Darmstadt, Germany). The membranes were blocked with 5% defatted milk for 2 hours at room temperature, followed by incubation with the primary antibody for 16 hours (1:1,000). Membranes were washed with Tris buffered saline Tween (TBST) and then incubated with a horse radish peroxidase (HRP) conjugated secondary antibody (1:2,000) for 2 hours at room temperature with agitation. The protein bands were analyzed using a chemiluminescence imaging analyzer. The primary antibodies used included the following: anti-BRD9 antibodies (Abcam, Cambridge, MA, USA; ab137245, 1:1,000), anti-GAPDH antibodies (Abcam, ab9485, 1:1,000), anti-E-cadherin antibodies (Cell Signaling Technology, Danvers, MA, USA; #14472, 1:1,000), anti-N-cadherin antibodies (Cell Signaling Technology, Danvers, RA, USA; #13116, 1:1,000), anti-Vimentin antibodies (Cell Signaling Technology, Danvers, RA, USA; #5741, 1:1,000), anti-MEK2 antibodies (Abcam, ab32517, 1:1,000), anti-ERK antibodies (Abcam, ab32537, 1:1,000), anti-p-MEK1/2 antibodies (Cell Signaling Technology, Danvers, RA, USA; #75262, 1:2,000), anti-p-ERK antibodies (Cell Signaling Technology, Danvers, RA, USA; #4370, 1:2,000), and anti-NCOA1 antibodies (Cell Signaling Technology, Danvers, RA, USA; #2191, 1:1,000). The secondary antibodies used were anti-mouse IgG (Promotor, Wuhan, China; 1:2,000) and anti-rabbit IgG (Promotor, 1:2,000). Each sample was repeated three times.
Cell proliferation assay
Cells in the logarithmic growth phase were inoculated in 96-well plates (5,000 cells/well), and each group was repeated 3 times. We observed the cultivation of the cells after 0, 24, 48, 72, and 96 hours. After 24 hours of incubation, 10 µL of CCK-8 reagent (Solarbio, Beijing, China) was added to each well and the cell proliferation rate was calculated by measuring the absorbance at 450 nm with an enzyme marker after 1 hour.
Colony formation assay
CRC cells were seeded into six-well plates at 1×103 cells/well using fresh culture medium incubated in a thermostatic incubator for 14 days. The culture medium was refreshed as appropriate. Cells were washed with phosphate buffered saline (PBS) 2–3 times, followed by methanol fixation for 5 minutes and staining with crystalline purple for 5 minutes. Cells were then washed with tap water and dried. The number of cell colonies was counted and photographed. Each group was repeated in triplicates.
TUNEL assay
Transfected cells were washed with PBS and fixed with 4% paraformaldehyde for 30 minutes. Subsequently, cells were incubated with PBS containing 0.3% Triton X-100 for 5 minutes at room temperature. After washing with PBS, 50 µL TUNEL test solution (Beyotime, Shanghai, China) was added and incubated in the dark at 37 ℃ for 60 minutes. Cells were observed under the fluorescence microscope (IX53, Olympus, Japan). Each group was repeated three times.
Flow cytometry
Apoptosis was evaluated by Annexin V-FITC/PI double staining flow cytometry. A total of 1×104 cells in logarithmic growth was resuspended in 400 µL phosphate buffer solution and incubated with 10 µL HRP-labeled Annexin V-FITC and 5 µL propidium iodide (PI; BioLegend, San Diego, CA, USA) for 30 minutes in the dark at room temperature. The apoptosis rate was detected by flow cytometry (Becton Dickinson, San Jose, CA, USA). Each group was repeated three times.
Transwell assay
Transfected cells were seeded into the upper chamber of the transwell and the lower chamber contained culture medium with 10% fetal bovine serum. After 48 hours culture, the chamber was removed and cells were fixed with polyformaldehyde and stained with 1% crystal violet for 5 minutes. The cells were observed and counted under a microscope at 200-fold magnification to obtain the average value number of migrating cells.
For invasion assays, the Matrigel gel was diluted with DMEM culture medium and spread on the upper chamber of the transwell containing an 8 µm polycarbonate insert. The lower contained culture medium supplemented with 10% fetal bovine serum. Transfected cells were seeded into the upper chamber of the transwell and cultured for 24 hours. After 24 hours of culture, the chamber was removed. The cells in the upper chamber of the transwell were gently wiped and the cells in the lower chamber were fixed with paraformaldehyde and dye with 1% crystal violet for 5 minutes. Cells passing through the filter membrane were observed under a microscope at 200-fold magnification and counted to obtain the average number of invading cell. Each group was repeated three times.
IHC
The collected CRC tissues and the adjacent tissues were fixed in 10% formalin and embedded in paraffin. Routine paraffin sections were prepared by baking the tissue slices in a constant temperature oven at 60 ℃ for 4 hours. Samples were maintained at 4 ℃ until immunohistochemical staining was performed. Slides were treated with H2O2 methanol solution for 15 minutes and incubated with the sealing solution for 10 minutes. The liquid was then removed and slides were incubated with a monoclonal antibody for 16 hours (1:100). After washing with PBS for 2 minutes, the slides were incubated with an anti-biotinylated secondary antibody for 10 minutes and washed with PBS for 2 minutes. Slides were then incubated with an HRP-labeled chain avidin for 10 minutes and washed with PBS for 2 minutes. DAB color development was performed, followed by washing with water, re-dyeing, dehydration, transparency, and sealing. Five visual fields were selected from each slice and examined under the microscope. Two experienced pathologists performed double-blinded identification and counted 100 cells in each visual field. The positive staining rate was calculated as the number of positive cells in the total number of cells.
In vivo xenograft assay
SW480 cell lines transfected with control vector and shBRD9 were routinely cultured. Five-week-old male BALB/c nude mice were housed in a specific pathogen free (SPF) animal laboratory because their immune system is completely ineffective, and any pathogenic agent can infect the BALB/c nude mice Animals were randomly divided into the vector group and the shBRD9 group (five mice in each group). Various methods were used to limit, match, randomize, stratify, standardize, and analyze the selection of subjects to control the confounding bias. Cells in logarithmic growth were used to prepare a cell suspension with a concentration of 1×106 cells/mL, which was then inoculated under the left axillary skin of the nude mice. Each nude mouse was subcutaneously inoculated with 0.2 mL of cells, and the tumor growth in the inoculated cells was observed every day after inoculation. Tumor formation was determined by tumor nodules and hard texture under the inoculation site. Three weeks later, the mice were euthanized, and the subcutaneous tumors were completely removed and measured. Collection of data through blind methods, collection of data on objective indicators, and an extensive collection of information to ensure the scientific attitude of the researchers were performed. All animal experiments were approved by the Experimental Animal Ethics Committee of the Chinese PLA General Hospital (S2022-095-01), in compliance with institutional guidelines for the care and use of animals. A protocol was prepared before the study without registration.
Bioinformatics analysis
The expression level of BRD9 was analyzed by using The Cancer Genome Atlas (TCGA) database (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga), which contained 698 CRC samples and para-tumor tissues. Gene set enrichment analysis (GSEA) was used to analyze the potential biological signaling pathways and functions of BRD9. Markedly enriched signaling pathways with determined using the enrichment score (NES), gene ratio, and P value. The correlation between BRD9 expression and the expression of other genes was analyzed using the Spearman’s correlation method.
Statistical analysis
The R language software (version 4.0.2) (https://www.r-project.org/) was used for statistical analyses. Continuous variables were defined as mean ± standard deviation (SD) or median (interquartile range). For continuous variables, t-test or a one-way analysis of variance (ANOVA) model was used for comparisons. The Spearman rank test was used to test correlations. Variables with a P<0.05 in the univariate analysis were subjected to multivariate analysis using a Cox proportional hazards model to determine independent prognostic factors. The Kaplan-Meier method and the log-rank test were used for survival analysis. All tests were two sided, and P value <0.05 was considered statistically significant.
Results
BRD9 is upregulated in CRC samples and cell lines
The mRNA expression of BRD9 was examined in five CRC cell lines and the colorectal mucosal epithelial cell line FHC. The mRNA expression of BRD9 was elevated in all CRC cell lines compared to FHC (Figure 1A). In addition, the level of BRD9 mRNA was upregulated in 28 out of 31 patients examined (Figure 1B). Furthermore, the protein level of BRD9 was upregulated in both CRC cell lines and typical patients (n=10; P<0.05; Figure 1C,1D). Additionally, the level of BRD9 mRNA in TCGA databases was higher in CRC samples compared to the normal groups (P<0.01; Figure 1E,1F). IHC staining also demonstrated that protein expression of BRD9 was shown different in CRC tissues compared to normal adjacent tissues (Figure 1G).
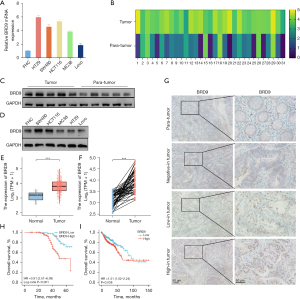
High BRD9 expression in CRC was correlated with poor survival
We analyzed the protein levels of 524 CRC patients using paraffin-embedded tissues (Table 1). Patients were separated into two groups according to the expression of BRD9, namely, the high BRD9 expression group (n=296), and the low BRD9 expression group (n=228). BRD9 was upregulated in most CRC tissues and Kaplan-Meier survival curves revealed that the overall survival (OS) was shorter in patients with high BRD9 expression (P<0.01; Figure 1H). These findings were consistent with the TCGA database (P<0.05; Figure 1I). Univariate and multivariate Cox regression analysis demonstrated that BRD9 was an independent risk factor for CRC patients [hazard ratio (HR): 3.04, 95% confidence interval (CI): 1.78–5.20; P<0.01; Table 2].
Table 1
| Variable | All patients (total n=524) | P value | |
|---|---|---|---|
| BRD9 low (n=228) | BRD9 high (n=296) | ||
| Age (years), mean ± SD | 69.2±7.8 | 70.1±10.8 | 0.33 |
| Sex (male), n [%] | 157 [69] | 210 [71] | 0.61 |
| MSI-H, n [%] | 22 [10] | 40 [14] | 0.18 |
| CEA (ng/mL), median (interquartile range) | 2.9 (1.5–14.2) | 1.8 (5.7–17.9) | <0.01 |
| Location, n [%] | 0.45 | ||
| Left colon | 47 [21] | 41 [14] | |
| Right colon | 80 [35] | 158 [83] | |
| Rectal | 101 [44] | 97 [33] | |
| Differentiation, n [%] | 0.12 | ||
| Well | 89 [39] | 138 [47] | |
| Moderate | 58 [25] | 66 [22] | |
| Poor | 81 [36] | 92 [31] | |
| Tumor size (mm), mean ± SD | 31.9±10.4 | 30.5±14.4 | 0.22 |
| T stage, n [%] | 0.09 | ||
| T1 | 8 [4] | 0 [0] | |
| T2 | 73 [32] | 95 [32] | |
| T3 | 103 [45] | 130 [44] | |
| T4 | 44 [19] | 71 [24] | |
| N stage, n [%] | <0.01 | ||
| N0 | 156 [68] | 69 [23] | |
| N1 | 51 [22] | 75 [23] | |
| N2 | 21 [9] | 152 [51] | |
| TNM, n [%] | <0.01 | ||
| I | 60 [26] | 6 [2] | |
| II | 96 [42] | 63 [21] | |
| III | 72 [32] | 227 [77] | |
BRD9, bromodomain-containing protein 9; CRC, colorectal cancer; SD,standard deviation ; MSI-H, microsatellite instability-high; CEA, carcinoembryonic antigen.
Table 2
| Variables | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | ||
| Age (<65/≥65 years) | 1.09 | 0.56–2.12 | 0.79 | ||||
| Sex (male/female) | 5.87 | 3.68–9.37 | <0.01 | 6.39 | 3.94–10.37 | <0.01 | |
| MSI-H (no/yes) | 1.19 | 0.63–2.19 | 0.58 | ||||
| CEA (<5/≥5 ng/mL) | 1.27 | 0.79–2.01 | 0.32 | ||||
| Location (colon/rectal) | 0.86 | 0.56–1.33 | 0.50 | ||||
| Differentiation (well-mod/poor) | 1.32 | 0.86–2.05 | 0.21 | ||||
| Depth of tumor (T1, T2, T3/T4) | 1.59 | 0.99–2.58 | 0.06 | ||||
| N stage (N0, NI/N2) | 4.38 | 2.79–6.84 | <0.01 | ||||
| TNM (I, II/III) | 7.48 | 3.97–14.10 | <0.01 | 4.54 | 2.25–9.13 | <0.01 | |
| BRD9 (low/high) | 4.12 | 2.61–6.64 | <0.01 | 3.04 | 1.78–5.20 | <0.01 | |
CRC, colorectal cancer; HR, hazard ratio; CI, confidence interval; MSI-H, microsatellite instability-high; CEA, carcinoembryonic antigen; TNM, tumor node metastasis; BRD9, bromodomain-containing protein 9; well-mod/poor, well-moderate/poor.
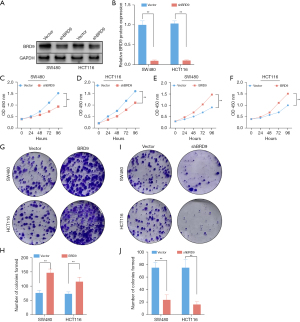
Downregulation of BRD9 suppressed the growth and viability of CRC cells
SW480 and HCT116 cell lines were transfected with a lentivirus carrying a shRNA against BRD9 (shBRD9). Western blot analysis confirmed that shBRD9 protein expression was significantly knocked down cells transfected with shBRD9 (P<0.01; Figure 2A,2B). CCK-8 assays revealed that BRD9 downregulation significantly inhibited CRC cell proliferation, while SW480 and HCT116 cell lines were transfected with a lentivirus carrying an overexpression against BRD9 (oeBRD9) markedly enhance cell proliferation (P<0.01; Figure 2C-2F). Colony formation assays demonstrated that the colony formation rate was significantly suppressed in cells with downregulated BRD9 expression, while the number of colon formation of SW480 and HCT116 cell lines was significantly enhanced when BRD9 expression was upregulated (P<0.01; Figure 2G-2J).
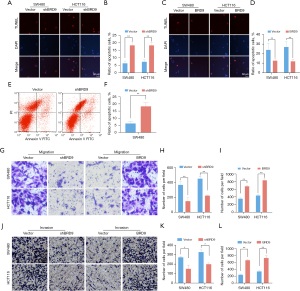
Effects of BRD9 on apoptosis, migration, and invasion of CRC cells in vitro
The TUNEL assay revealed that downregulated BRD9 expression enhanced the apoptosis of CRC cells, while overexpression of BRD9 decreased the percentage of apoptotic cells in SW480 and HCT116 cells (P<0.01; Figure 3A-3D). Flow cytometry confirmed the enhanced numbers of apoptotic cells in SW480 cells transfected with shBRD9 (P<0.01; Figure 3E,3F). Migration of SW480 and HCT116 cells were detected by transwell assay (Figure 3G) and cells were counted (P<0.01; Figure 3H,3I). Invasion of SW480 and HCT116 cells were detected by transwell assay (Figure 3J) and cells were counted (P<0.05; Figure 3K,3L)
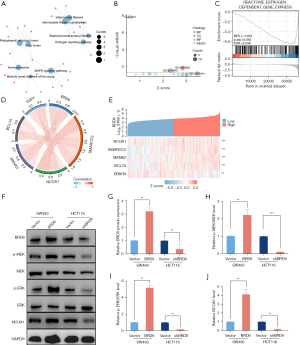
Inactivation of the estrogen pathway resulted in BRD9 knockdown induced growth suppression in CRC cells BRD9 regulated the estrogen signaling pathway
Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) enrichment datasets were analyzed to identify the signaling pathways associated with BRD9. The KEGG pathways associated with BRD9 and tumor proliferation included the “MAPK signaling pathway” and the “estrogen signaling pathway” (Figure 4A). GO enrichment analysis indicated that the genes interacting with BRD9 were related to signaling pathways or biological functions of cells, including ligand activity chromatin remodeling, hormone activity, maturity-onset diabetes of the young, and Staphylococcus aureus infection (Figure 4B). To analyze which pathways were mediated by BRD9, GSEA analysis was performed on TCGA CRC database (Figure 4C). The results demonstrated that the level of BRD9 was negatively related to estrogen-dependent gene expression. Moreover, BRD9 expression was positively correlated with nuclear receptor coactivator 1 (NCOA1; Figure 4D,4E). The effect of BRD9 overexpression and silencing on BRD9 protein expression in SW480 and HCT116 cells was confirmed by Western blots (Figure 4F,4G). BRD9 overexpression increased the ratio of phosphorylated (p)-MEK/MEK (P<0.01; Figure 4F,4H), p-ERK/ERK (P<0.01; Figure 4F,4I), and NCOA1 (P<0.01; Figure 4F,4J) in SW480 cells. Meanwhile, downregulation of BRD9 reduced the ratio of p-MEK/MEK (P<0.01; Figure 4F,4H), p-ERK/ERK (P<0.01; Figure 4F,4I), and the levels of NCOA1 (P<0.01; Figure 4F,4J).
Knockdown of BRD9 suppressed EMT via loss of function in the estrogen pathway in CRC cells
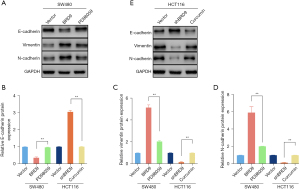
Numerous studies have revealed that the estrogen pathway might play a significant role in contributing to EMT in CRC. Therefore, the role of the BRD9/estrogen pathway on EMT in CRC was examined. To regulate the activity of the estrogen pathway, PD98059 (a MEK inhibitor) and curcumin (an ERK activator) were used. Western blot analyses demonstrated that overexpression of BRD9 notably decreased the protein expression of E-cadherin (P<0.01; Figure 5A,5B) and promoted the expression of vimentin (P<0.01; Figure 5A,5C) and N-cadherin (P<0.01; Figure 5A,5D). BRD9-induced EMT was markedly decreased by PD98059 (P<0.01; Figure 5A,5B-5D). Conversely knockdown of BRD9 notably increased the levels of E-cadherin (P<0.01; Figure 5E,5B) and decreased vimentin (P<0.01; Figure 5E,5C) and N-cadherin (P<0.01; Figure 5E,5D) expression. However, the inhibitory effects of shBRD9 on EMT were reserved by curcumin (Figure 5E,5B-5D). These results suggested that downregulation of BRD9 suppressed EMT via loss of function in the estrogen pathway in CRC cells.
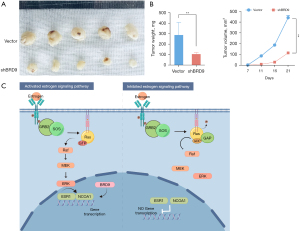
Knockdown of BRD9 inhibits tumorigenesis in vivo
Finally, BALB/c nude mice were used to determine the influence of BRD9 on tumorigenesis. BALB/c nude mice were subcutaneously inoculated with SW480 cells that had been transfected with shBRD9 or control vector. Tumors were dissected from the mice after three weeks (Figure 6A). The volume and weight of the tumors were markedly decreased in the shBRD9 group compared with the vector group (P<0.01; Figure 6B), suggesting that BRD9 knockdown markedly suppressed tumorigenesis in vivo for CRC cells. This study revealed that BRD9 facilitates CRC progression by regulating NCOA1 expression (Figure 6C).
Discussion
This investigation demonstrated that BRD9 was overexpression in both CRC tissues and cell lines. Kaplan-Merier survival curves showed that CRC patients with high expression of BRD9 were associated with a shorter OS compared to patients with low BRD9 expression. Univariate and multivariate analyses suggested that BRD9 might be an independent risk factor for CRC patients. Additionally, upregulating BRD9 enhanced tumorigenesis and growth in CRC. Furthermore, our study suggested that knockdown of BRD9 suppressed EMT via loss of function in the estrogen pathway of CRC cells. In conclusion, tumorigenesis was suppressed after downregulating BRD9 expression in CRC cells.
As an essential epigenetic regulatory factor, numerous genes, such as MYC, are regulated by BRD- containing protein (17). Additionally, BRD-containing proteins participate in various life processes (18) and has been shown to play a crucial role in tumor occurrence and development (19,20). BRD-containing proteins recently been identified from the SWI/SNF complex. However, the composition of SWI/SNF varies with cell type and tissues (21), and bromodomain-containing subunits, such as BRD9, have potentially targetable chromatin reader domains. At present, the complex function of the BRD9 protein is not well understood. However, BRD9 has been reported to play a vital role in numerous tumors. Del Gaudio et al. suggested that BRD9 may be a potential therapeutic target via the BRD9-STAT5 axis and is critical for leukemia maintenance (22). Huang et al. indicated that inhibition of BRD9 repressed squamous cell lung cancer tumorigenesis by regulating c-myc expression (23). Mu et al. showed that BRD9 plays a vital role in the progression of gastrointestinal stromal tumors (GISTs), and inhibition of BRD9 may be a novel therapeutic strategy against GISTs, either alone or in combination with imatinib (24). There are a variety of commercial inhibitors which have been shown to be effective at inhibiting malignant tumor phenotypes (25). BRD9 protein has many corresponding inhibitors (26,27), including I-BRD9 which has reasonable specificity and a noticeable inhibitory effect on BRD9 protein activity (28). Moreover, the deep bromodomain pocket can be targeted with small-molecules (17).
The estrogen pathway plays a critical role in the development and progression of CRC and warrants further investigation, especially in relation to targeted therapeutic strategies. Jiang et al. demonstrated that injection of E2 supplements in mice inhibited the proliferation of colon cancer cells. E2 may hinder the growth of MC38 tumors by inhibiting the proportion of myeloid-derived suppressor cells rather than by inhibiting the proliferation or promoting the apoptosis of MC38 cells (29). In an animal model of inflammation-associated CRC, enriched formulation exerted a significant chemo-prevention due to inflammation reduction, epithelial turnover/apoptosis increase, and estrogen receptor beta stimulation (30). Our study suggested that BRD9 is negatively correlated with estrogen, resulting in poor survival for CRC patients.
Colorectal immune regulation mainly involves colorectal mucosal epithelial cells, intestinal intraepithelial lymphocytes, and innate lymphocytes (including dendritic cells, intestinal T cells, and plasma cells). Moreover, dendritic cells, intestinal T cells, and plasma cells play an important role in the tumor microenvironment. In addition, BRD9 has been reported to be related to the tumor microenvironment, immune cell infiltration, and immune checkpoints, and was verified to be significantly associated with prognosis and tumor mutation burden in clear cell renal carcinoma (31). Thus, we mean to investigate the relationship between BRD9 and the immune microenvironment in colorectal cancer and develop new drug treatment methods in the future study.
Growing evidence supports the idea that the estrogen receptor (ER) promotes EMT in many cancers. Estrogen significantly downregulates miR-765 levels and further regulates Ki-67 and multiple EMT-related molecules in uterine corpus endometrial cancer patients (32). ZEB1 modulates ERα-mediated transcription induced by EMT via estrogen or cAMP signaling in breast cancer cells (33). EMT of melanoma cells induced by extracellular acidosis correlates with the expression of Erβ (34). Circ_0000799 promotes proliferation and invasion in colorectal cancer and epithelial-mesenchymal transition (35). PBX1 interacted with ERs and is required for ER function, and overexpression of PBX1 promoted the cell EMT process in bladder cancer cells (36). ERα downregulation inhibited the proliferation, migration, and invasion of gastric cancer, via regulating the expression of p53, p21, p27, and E-cadherin (14). Our study supports a role for the estrogen pathway in facilitating EMT in CRC. Since the occurrence and development of EMT are related to the metastasis of tumors, and the liver is the most frequent site of metastases and dominates the length of survival for colorectal cancer, while the study about the mechanism of colorectal cancer with liver metastasis is rare, further studies using in vivo colorectal cancer models of liver metastasis will reveal the role of BRD9 in colorectal cancer with liver metastasis.
Cancer stem cells (CSCs) and EMT promote the progression of CRC patients with liver metastasis. Moreover, EMT could induce the formation of CSCs and increase drug resistance. While, the specific molecular mechanisms and therapeutic resistance mechanisms between BRD9, EMT, and CSCs are still unclear. Activation of JAK/STAT3 signaling pathways induces the EMT, and results in increasing tumorigenicity and metastasis, promoting CSCs transformation, and drug resistance for CRC (37). The therapeutic strategies targeting the biomarkers of EMT are expected to overcome the drug resistance, cancer progression, and ultimately recurrence of CSCs in CRC. Our study has shown that BRD9 promotes the occurrence of EMT, thus we plan to explore the potential relationships between BRD9, EMT, and CRCs in the near future.
Conclusions
In conclusion, BRD9 might be an independent risk factor affecting the occurrence of CRC, and the BRD9/estrogen pathway is closely related to the proliferation, invasion, and apoptosis of CRC tissues and cell lines, suggesting that BRD9 might be a potential target for the treatment of postmenopausal women with CRC.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR and ARRIVE reporting checklists. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-271/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-271/dss
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-271/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-271/coif). The authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Hendifar A, Yang D, Lenz F, et al. Gender disparities in metastatic colorectal cancer survival. Clin Cancer Res 2009;15:6391-7. [Crossref] [PubMed]
- Delattre JF, Selcen Oguz Erdogan A, Cohen R, et al. A comprehensive overview of tumour deposits in colorectal cancer: Towards a next TNM classification. Cancer Treat Rev 2022;103:102325. [Crossref] [PubMed]
- Sanchez R, Zhou MM. The role of human bromodomains in chromatin biology and gene transcription. Curr Opin Drug Discov Devel 2009;12:659-65. [PubMed]
- Fujisawa T, Filippakopoulos P. Functions of bromodomain-containing proteins and their roles in homeostasis and cancer. Nat Rev Mol Cell Biol 2017;18:246-62. [Crossref] [PubMed]
- Ali MM, Ashraf S, Nure-E-Alam M, et al. Identification of Selective BRD9 Inhibitor via Integrated Computational Approach. Int J Mol Sci 2022;23:13513. [Crossref] [PubMed]
- Wang X, Wang S, Troisi EC, et al. BRD9 defines a SWI/SNF sub-complex and constitutes a specific vulnerability in malignant rhabdoid tumors. Nat Commun 2019;10:1881. [Crossref] [PubMed]
- Zhang C, Chen L, Lou W, et al. Aberrant activation of m6A demethylase FTO renders HIF2α(low/-) clear cell renal cell carcinoma sensitive to BRD9 inhibitors. Sci Transl Med 2021;13:eabf6045. [Crossref] [PubMed]
- Weisberg E, Chowdhury B, Meng C, et al. BRD9 degraders as chemosensitizers in acute leukemia and multiple myeloma. Blood Cancer J 2022;12:110. [Crossref] [PubMed]
- Takaku M, Grimm SA, Shimbo T, et al. GATA3-dependent cellular reprogramming requires activation-domain dependent recruitment of a chromatin remodeler. Genome Biol 2016;17:36. [Crossref] [PubMed]
- Barma N, Stone TC, Carmona Echeverria LM, et al. Exploring the Value of BRD9 as a Biomarker, Therapeutic Target and Co-Target in Prostate Cancer. Biomolecules 2021;11:1794. [Crossref] [PubMed]
- Zhang N, Ng AS, Cai S, et al. Novel therapeutic strategies: targeting epithelial-mesenchymal transition in colorectal cancer. Lancet Oncol 2021;22:e358-68. [Crossref] [PubMed]
- Barzi A, Lenz AM, Labonte MJ, et al. Molecular pathways: Estrogen pathway in colorectal cancer. Clin Cancer Res 2013;19:5842-8. [Crossref] [PubMed]
- Tang W, Liu R, Yan Y, et al. Expression of estrogen receptors and androgen receptor and their clinical significance in gastric cancer. Oncotarget 2017;8:40765-77. [Crossref] [PubMed]
- Kapoor S, Damiani E, Wang S, et al. BRD9 Inhibition by Natural Polyphenols Targets DNA Damage/Repair and Apoptosis in Human Colon Cancer Cells. Nutrients 2022;14:4317. [Crossref] [PubMed]
- Zhu Q, Gu X, Wei W, et al. BRD9 is an essential regulator of glycolysis that creates an epigenetic vulnerability in colon adenocarcinoma. Cancer Med 2023;12:1572-87. [Crossref] [PubMed]
- Hohmann AF, Martin LJ, Minder JL, et al. Sensitivity and engineered resistance of myeloid leukemia cells to BRD9 inhibition. Nat Chem Biol 2016;12:672-9. [Crossref] [PubMed]
- Banerjee C, Archin N, Michaels D, et al. BET bromodomain inhibition as a novel strategy for reactivation of HIV-1. J Leukoc Biol 2012;92:1147-54. [Crossref] [PubMed]
- Urbanucci A, Barfeld SJ, Kytölä V, et al. Androgen Receptor Deregulation Drives Bromodomain-Mediated Chromatin Alterations in Prostate Cancer. Cell Rep 2017;19:2045-59. [Crossref] [PubMed]
- Wang CY, Filippakopoulos P. Beating the odds: BETs in disease. Trends Biochem Sci 2015;40:468-79. [Crossref] [PubMed]
- Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell 2009;136:200-6. [Crossref] [PubMed]
- Del Gaudio N, Di Costanzo A, Liu NQ, et al. BRD9 binds cell type-specific chromatin regions regulating leukemic cell survival via STAT5 inhibition. Cell Death Dis 2019;10:338. [Crossref] [PubMed]
- Huang H, Wang Y, Li Q, et al. miR-140-3p functions as a tumor suppressor in squamous cell lung cancer by regulating BRD9. Cancer Lett 2019;446:81-9. [Crossref] [PubMed]
- Mu J, Sun X, Zhao Z, et al. BRD9 inhibition promotes PUMA-dependent apoptosis and augments the effect of imatinib in gastrointestinal stromal tumors. Cell Death Dis 2021;12:962. [Crossref] [PubMed]
- Krämer KF, Moreno N, Frühwald MC, et al. BRD9 Inhibition, Alone or in Combination with Cytostatic Compounds as a Therapeutic Approach in Rhabdoid Tumors. Int J Mol Sci 2017;18:1537. [Crossref] [PubMed]
- Clark PG, Vieira LC, Tallant C, et al. LP99: Discovery and Synthesis of the First Selective BRD7/9 Bromodomain Inhibitor. Angew Chem Weinheim Bergstr Ger 2015;127:6315-9. [Crossref] [PubMed]
- Martin LJ, Koegl M, Bader G, et al. Structure-Based Design of an in Vivo Active Selective BRD9 Inhibitor. J Med Chem 2016;59:4462-75. [Crossref] [PubMed]
- Theodoulou NH, Bamborough P, Bannister AJ, et al. Discovery of I-BRD9, a Selective Cell Active Chemical Probe for Bromodomain Containing Protein 9 Inhibition. J Med Chem 2016;59:1425-39. [Crossref] [PubMed]
- Jiang L, Fei H, Yang A, et al. Estrogen inhibits the growth of colon cancer in mice through reversing extracellular vesicle-mediated immunosuppressive tumor microenvironment. Cancer Lett 2021;520:332-43. [Crossref] [PubMed]
- Girardi B, Principi M, Pricci M, et al. Chemoprevention of inflammation-related colorectal cancer by silymarin-, acetyl-11-keto-beta-boswellic acid-, curcumin- and maltodextrin-enriched dietetic formulation in animal model. Carcinogenesis 2018;39:1274-82. [Crossref] [PubMed]
- Deng Z, Zhang Y, Zhu Y, et al. BRD9 Inhibition Attenuates Matrix Degradation and Pyroptosis in Nucleus Pulposus by Modulating the NOX1/ROS/NF-κB axis. Inflammation 2023; Epub ahead of print. [Crossref] [PubMed]
- Zhou WJ, Zhang J, Xie F, et al. CD45RO(-)CD8(+) T cell-derived exosomes restrict estrogen-driven endometrial cancer development via the ERβ/miR-765/PLP2/Notch axis. Theranostics 2021;11:5330-45. [Crossref] [PubMed]
- Mohammadi Ghahhari N, Sznurkowska MK, Hulo N, et al. Cooperative interaction between ERα and the EMT-inducer ZEB1 reprograms breast cancer cells for bone metastasis. Nat Commun 2022;13:2104. [Crossref] [PubMed]
- Peppicelli S, Ruzzolini J, Lulli M, et al. Extracellular Acidosis Differentially Regulates Estrogen Receptor β-Dependent EMT Reprogramming in Female and Male Melanoma Cells. Int J Mol Sci 2022;23:15374. [Crossref] [PubMed]
- Geng CH, Zhang XS, He M, et al. Circ_0000799 promotes proliferation and invasion in colorectal cancer and epithelial-mesenchymal transition. J Gastrointest Oncol 2022;13:3090-9. [Crossref] [PubMed]
- Zhao Y, Che J, Tian A, et al. PBX1 Participates in Estrogen-mediated Bladder Cancer Progression and Chemo-resistance Affecting Estrogen Receptors. Curr Cancer Drug Targets 2022;22:757-70. [Crossref] [PubMed]
- Jin W. Role of JAK/STAT3 Signaling in the Regulation of Metastasis, the Transition of Cancer Stem Cells, and Chemoresistance of Cancer by Epithelial-Mesenchymal Transition. Cells 2020;9:217. [Crossref] [PubMed]
(English Language Editor: J. Teoh)


