
Mutant KRAS inhibitors enter the scene of precision therapeutics for pancreatic cancer
One of the most important programs within the Linda T. and John A. Mellowes Center for Genomic Sciences and Precision Medicine focuses on better understanding RASopathies, which are diseases caused by germline KRAS mutations. Similarly important, this program brings discoveries and innovation to comprehend the pathobiology of somatic KRAS mutations in cancer, particularly pancreatic ductal adenocarcinoma (PDAC). Indeed, in the last 4 decades, we have studied the molecular basis of PDAC. In particular, we have addressed how epigenomic pathways work downstream of KRAS to give rise to cancer initiation and progression. Recently, we developed data science and experimental methodologies to examine these disease-causing mutations, revealing the biophysical mechanisms displayed by 935 distinct missense mutations that affect the RAS family of genes (1,2), and including 188 distinct KRAS missense mutations already observed across RASopathies and cancers. Thus, with extensive experience working on genomics and epigenomics of pancreatic cancer, one of the most aggressive and devastating oncological diseases, we are fortunate to witness the discovery of MRTX1133. This oral inhibitor is showing promise as a therapeutic intervention targeting the KRASG12D mutation, as discussed in the recent article “A small molecule with big impact: MRTX1133 targets the KRASG12D mutation in pancreatic cancer”, written by Wei et al. [2024]. The work we describe and illustrate at atomic resolution in this article supports the observations discussed by our esteemed colleagues and pancreatic cancer experts from MD Anderson (3). The invention of KRASG12D inhibitors such as MRTX1133 marks the start of a new precision oncogenic therapeutic approach for pancreatic cancer. Moreover, we expand the discussion by addressing the necessity and current studies for targeted therapy for the less common and less-investigated subtypes of pancreatic cancer. This latest innovation in KRAS therapeutics brought to mind a similar work shedding light on the first publication using KRAS as a marker for pancreatic cancer and the challenges posed at that time (4). In almost three decades, the narrative of KRAS has shifted from a marker to now a target for the therapeutics of pancreatic cancer.
Pancreatic cancer and KRAS mutations
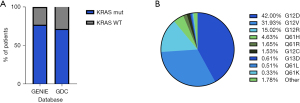
Pancreatic cancer remains one of the most lethal cancers, with an age-standardized rate (ASR) incidence of 4.7 and a mortality ASR of 4.2, ranked 12th and 6th, respectively, of all cancers (5). In the last 30 years, the pancreatic cancer 5-year survival rate has only increased to 13%. Hence, the clear need for better treatments (6). Currently, predictive tests for treatment response or prognosis are lacking. Therefore, clinicians rely on the general condition of the patient and the stage of the disease when selecting treatment options. In particular, PDAC is the predominant type, generally developing from low-grade pancreatic intraepithelial neoplasia (PanINs), to high-grade PanINs, and ultimately invasive adenocarcinoma (7). Initiation and progressions of PanIN lesions coincide with genetic and epigenetic mutations, with activating KRAS mutations being nearly ubiquitous in low-grade PanINs (8-10). KRAS mutations are often cited as being involved in 90% of pancreatic cancer cases. In reality this number tends to be more variable (11). For instance, the American Association for Cancer Research (AACR) Project Genomics Evidence Neoplasia Information Exchange (GENIE) reports a prevalence of 77.5%, and the NIH Genomic Data Commons (GDC) cancer portal reported a lower figure of 71.7% for single-base missense mutations in KRAS (12,13). Regardless of the source, they are consistently in agreement that KRAS mutations are the common denominator across pancreatic cancer (Figure 1A). Further insights from the GENIE Consortium v15.1 emphasized the heterogeneity in subtypes of KRAS mutations found in pancreatic cancer. KRAS single substitutions at the G12 residue constitute over 90% of KRAS-mutated pancreatic patients, largely driven by G12D, G12V, and G12R. Furthermore, we observe other notable subtypes, such as mutations at Q61 and G13 (Figure 1B). Thus, the recent successes of small molecule compounds targeting KRAS mutants have marked the onset of promising precision oncogenic therapeutic strategies to improve the treatment options for this fatal disease.
Discovery of KRASG12D inhibitor MRTX1133
The family of RAS proteins has been the target of cancer-related therapies for over 40 years and was often referred to as ‘undruggable’ due to the lack of classical drug binding sites. Excitingly, a turning point occurred in May 2021 when sotorasib, the first KRAS inhibitor targeting the G12C mutant, was granted FDA approval for the treatment of adults with advanced non-small cell lung cancer (NSCLC). Later, similar approval was granted for adagrasib in 2022. This discovery resulted in an eruption of drug development of small molecule inhibitors targeting G12C, entering into clinical trials testing variations potency and potential synergies with other drugs (14). Inspired by the success of targeting KRASG12C with adagrasib, Mirati Therapeutics has started their evaluation of the KRASG12D inhibitor MRTX1133, which has also entered clinical trials (NCT05737706) and is anticipated to conclude by August 2026 (15,16). As accurately described by Wei et al. [2024] recent experimentations are worth laudatory comments. For instance, utilizing preclinical mutant KRASG12D xenograft models from PDAC cell lines or primary patient-derived tumors, MRTX1133 observed significant anti-tumor activity, with tumor regression observed in 8 of the 11 PDAC models investigated. Furthermore, differential expression of key cancer cell-proliferation pathways was identified in xenografts treated with MRTX1133, these included KRAS signaling, MYC targets, mTORC, E2F targets, and G2M checkpoint. In addition, CRISPR screening guided the discovery of genes for developing combination therapy of MRTX1133 with human epidermal growth factor receptor (HER) family inhibitors afatinib and cetuximab, as well as the PI3Kα inhibitor BYL-719, resulting in synergistic effects in PDAC cells (15). Recently, Kumarasamy et al. [2024] reported that inhibition of KRASG12D by MRTX1133 modulated the tumor microenvironment by boosting the IFNγ signaling, and antigen presentation promoting an influx of cancer-fighting CD8+ Tells into the tumor microenvironment (17). Thus, in the era of immunotherapies, this information acquires significant biomedical relevance.
Despite the promising results for KRASG12D cancer therapeutics, it is important to acknowledge potential challenges as we navigate this new era of inhibitors. Firstly, like any new cancer drug discovery, it poses major hurdles in the development of resistance, with one potential mechanism including copy number variance (CNV). To highlight this potential issue, we examined the GDC Cancer Portal database confirming that 20.46% of mutant KRAS pancreatic cancer patients exhibit gains in CNV. Another mechanism of resistance could arise from duplicate mutations. Again, our examination of the GENIE consortium v15.1, 3.1% of KRAS mutant pancreatic cancer patients carry a secondary mutation in other key residues, suggesting that a single specific inhibitor, alone, may not be as effective. A third challenge to be overcome is the remaining type of mutations. While KRASG12D is the major mutation in pancreatic cancer, it still covers only 42% of patients. Thus, there is a need for further research for inhibitors to target the other major G12 mutations, G12R and G12V, as well as the rare subtype mutations at other residues, such as the Q61 and G13 mutations. Additionally, direct inhibition of each mutated KRAS protein may not have the same effect in each body tissue due to the unique combination of effector and downstream signaling molecules present in each body tissue. Lastly, there has been an increase in research aimed at counteracting the development of activation in KRAS-related pathways, particularly in treatment-induced resistance to mutant KRAS inhibition. Specifically, alterations in receptor tyrosine kinases (RTK), mitogen-activated protein kinase (MAPK), or phosphoinositide 3-kinase (PI3K) pathways were observed in solid tumors treated with divarasib, a KRAS G12C inhibitor (18). One approach to addressing this mechanism of resistance is the use of combination therapy. For instance, a Phase 3 trial utilized the EGFR inhibitor panitumumab in combination with the KRAS G12C inhibitor sotorasib in patients with chemorefractory metastatic colorectal cancer. This study found that the combination treatment resulted in longer progression-free survival compared to standard treatment (19). More research is needed to understand better how each mutation KRAS couples to the membrane signaling complex, leading to distinct and heterogeneous signatures in different pathological contexts. Understanding the context of the distinct and heterogeneous signatures will facilitate the development of additional combinational therapies. This approach aims to enhance mutant KRAS-targeted therapy and overcome current challenges. Despite these challenges that currently remain, the discovery of mutant KRAS inhibitors has the potential to transform the landscape of PDAC therapy through precision medicine.
Emerging drugs and future directions
In this section, we highlight the commentary by Wei et al. [2024] and further expand this information to describe challenges and precision therapeutics that are currently in clinical trials. We are optimistic that combined, this data will be of significant interest to the field. Indeed, we underscore the fact that the mutant KRAS inhibitor field has rapidly evolved since the proverbial “opening of Pandora’s box” in 2021. Preliminary investigations, such as clinical trials and abstracts submitted to national societies, underscore how the field is already tackling the challenges of mutant KRAS therapeutics. For instance, an abstract submitted to the AACR conference for targeting RAS, investigated CNV and mutant allele fractions. The authors concluded that both factors may play important roles in resistance development and may potentially contribute to criteria selection for treatment (20). Additionally, a recent poster presented at the AACR annual meeting found substantial anti-tumor activity in xenografts treated with RMC-5127, a compound for a mutant-selective tri-complex inhibitor of the GTP-bound active form of KRAS G12V (21). A quick search into clinicaltrials.gov for mutant subtypes found several actively recruiting studies for KRASG12C [70], G12D [17], G12R [4], G12V [11] and G13D [3], but there were no studies active for the Q61 residue. Table 1 summarizes currently active clinical trials recruiting participants for the major KRAS G12 subtypes (G12D, G12R, and G12V) these trials include a mix of small molecule inhibitors, vaccines, and T-cell receptors (TCR) therapies targeting either a single mutation or Pan-mutant KRAS.
Table 1
| Drug | Target | NCT number | Type |
|---|---|---|---|
| TSN1611 | G12D | NCT06385925 | Small molecule |
| MRTX1133 | G12D | NCT05737706 | Small molecule |
| HRS-4642 | G12D | NCT05533463 | Small molecule/combination |
| Anti-KRASG12D mTCR | G12D | NCT03745326 | mTCR |
| NT-112 | G12D | NCT06218914 | TCR |
| ASP4396 | G12D | NCT06364696 | Small molecule |
| ASP3082 | G12D | NCT05382559 | Small molecule |
| RMC-9805 | G12D | NCT06040541 | Small molecule |
| CAR-DC | G12D; G12C; G12V | NCT05631899 | Vaccine |
| Mutant KRAS-peptide vaccine | G12C; G12V; G12D; G12A; G13D; G12R | NCT05254184 | Vaccine/combination |
| QTX3034 | G12D | NCT06227377 | Small molecule/combination |
| ELI-002 7P | G12D; G12R; G12V; G12A; G12C; G12S; G13D | NCT05726864 | Vaccine |
| BDTX-4933-101 | G12D; G12V; G13C | NCT05786924 | Small molecule |
| INCB161734 | G12D | NCT06179160 | Small molecule/combination |
| KISIMA-02 | G12D; G12V | NCT05846516 | Vaccine/combination |
| HLA-A*11:01 | G12V | NCT04146298 | TCR |
| Anti-KRAS G12V mTCR PBL | G12V | NCT03190941 | mTCR |
| FHA11KRASG12V-TCR IV | G12V | NCT06043713 | TCR |
| AFNT-211 | G12V | NCT06105021 | TCR |
This table summarizes ongoing clinical trials investigating drugs targeting mutations of G12 hotspot mutants in KRAS. Each entry includes the drug name, the specific KRAS mutation targeted (e.g., G12D, G12V, G12R), the NCT Number, and the drug type classified by the type of drug (e.g., TCR, small molecule, vaccine). Use the NCT Number to find detailed information about each trial on clinicaltrials.gov. TCR, T-cell receptor; mTCR, murine T-cell receptor; NCT, National Clinical Trial.
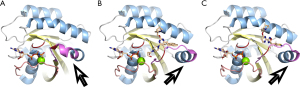
Moving forward, there are promising KRAS pan-mutant inhibitors not currently in clinical trials that hold relevance for PDAC. Using experimentally resolved wild-type (WT) KRAS and mutant KRAS, we demonstrate structural changes caused by the pan-mutant KRAS inhibitors (Figure 2A). YK-8S, a G12C/G12D dual KRAS inhibitor which selectively binds G12C mutations when bound to GDP as well as target G12D mutations when bound to guanosine 5'-[β,γ-imido] triphosphate (GppNHp), while demonstrating low modification of WT or other mutants (G12R, G13D, Q61R, and Q61K) (22). YK-8S binds above switch-II, partly occluding the space usually occupied by the third phosphate group of ATP and stabilizing switch-II in a more extended conformation (Figure 2B). Another inhibitor, BI-2865, stands out as a particularly exciting compound for the future of PDAC research. BI-2865 has been found to broadly target several KRAS mutants, and to a lesser extent WT KRAS, by blocking nucleotide exchange and preventing activation (23). BI-2865 binding results in the same type of extended switch-II conformation as YK-8S without occluding the third triphosphate position (Figure 2C). Interestingly, BI-2865 binding to the side of the p-loop and triphosphate grove may explain its efficacy against multiple p-loop mutations. As Stable switch conformations are required for KRAS to bind to its downstream effectors, the stabilizing extended Switch conformations (emphasized by the arrows in Figure 2) caused by these pan-mutant KRAS inhibitors likely prevent molecular coupling, leading to cessation of signaling through this axis. Further research into inhibitors targeting such a broad range of residues mutated in KRAS, from the G12 hotspot to the rare Q61, would indeed represent a significant advancement in enhancing therapeutic options for all mutant KRAS PDAC subtypes. Although the Q61 subtypes are not highly prevalent in PDAC, the development of pan-mutant KRAS drugs capable of targeting other hotspots such as G13 and Q61 would not only benefit the rare PDAC subtypes but also a wide range of other cancers, including melanoma, thyroid, urinary, and hematopoietic cancers (24).
In conclusion, the original article “A small molecule with big impact: MRTX1133 targets the KRASG12D mutation in pancreatic cancer”, written by Wei et al. [2024], serves as a useful guide to harnessing the potential of new KRAS inhibitors for treating PDAC. Furthermore, we alter to the fact that the increasing studies from our laboratory and others on the different pathobiological properties of different KRAS mutations are facilitating the discovery of mutant KRAS inhibitors, representing an exciting and promising area of precision oncogenic therapeutics. The continued research into mutant KRAS inhibitors is essential to generate additional FDA-approved inhibitors to target not only the common subtypes but also the rare subtypes in PDAC. This advancement holds immense potential to revolutionize the treatment landscape for PDAC, offering hope for improved outcomes and quality of life for patients through precision medicine.
Acknowledgments
Funding: This work was supported by
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Journal of Gastrointestinal Oncology. The article has undergone external peer review.
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-326/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-326/coif). The authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Tripathi S, Dsouza NR, Mathison AJ, et al. Enhanced interpretation of 935 hotspot and non-hotspot RAS variants using evidence-based structural bioinformatics. Comput Struct Biotechnol J 2021;20:117-27. [Crossref] [PubMed]
- Tripathi S, Dsouza NR, Urrutia R, et al. Structural bioinformatics enhances mechanistic interpretation of genomic variation, demonstrated through the analyses of 935 distinct RAS family mutations. Bioinformatics 2021;37:1367-75. [Crossref] [PubMed]
- Wei D, Wang L, Zuo X, et al. A Small Molecule with Big Impact: MRTX1133 Targets the KRASG12D Mutation in Pancreatic Cancer. Clin Cancer Res 2024;30:655-62. [Crossref] [PubMed]
- Urrutia R, DiMagno EP. Genetic markers: the key to early diagnosis and improved survival in pancreatic cancer? Gastroenterology 1996;110:306-10. [Crossref] [PubMed]
- Ferlay J, Ervik M, Lam F, et al. Global Cancer Observatory: Cancer Today. Lyon, France: International Agency for Research on Cancer.2024. Available online: https://gco.iarc.who.int/today. Accessed April 28, 2024.
- American Cancer Society. Cancer Facts & Figures 2024. Atlanta: American Cancer Society; 2024.
- Basturk O, Adsay NV. Early Cancerous Lesions of the Pancreas and Ampulla: Current Concepts and Challenges. Gastroenterol Clin North Am 2024;53:57-84. [Crossref] [PubMed]
- Burdziak C, Alonso-Curbelo D, Walle T, et al. Epigenetic plasticity cooperates with cell-cell interactions to direct pancreatic tumorigenesis. Science 2023;380:eadd5327. [Crossref] [PubMed]
- Hezel AF, Kimmelman AC, Stanger BZ, et al. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev 2006;20:1218-49. [Crossref] [PubMed]
- Lomberk G, Dusetti N, Iovanna J, et al. Emerging epigenomic landscapes of pancreatic cancer in the era of precision medicine. Nat Commun 2019;10:3875. [Crossref] [PubMed]
- Huang L, Guo Z, Wang F, et al. KRAS mutation: from undruggable to druggable in cancer. Signal Transduct Target Ther 2021;6:386. [Crossref] [PubMed]
- AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov 2017;7:818-31. [Crossref] [PubMed]
- Grossman RL, Heath AP, Ferretti V, et al. Toward a Shared Vision for Cancer Genomic Data. N Engl J Med 2016;375:1109-12. [Crossref] [PubMed]
- Batrash F, Kutmah M, Zhang J. The current landscape of using direct inhibitors to target KRAS(G12C)-mutated NSCLC. Exp Hematol Oncol 2023;12:93. [Crossref] [PubMed]
- Hallin J, Bowcut V, Calinisan A, et al. Anti-tumor efficacy of a potent and selective non-covalent KRAS(G12D) inhibitor. Nat Med 2022;28:2171-82. [Crossref] [PubMed]
- Wang X, Allen S, Blake JF, et al. Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS(G12D) Inhibitor. J Med Chem 2022;65:3123-33. [Crossref] [PubMed]
- Kumarasamy V, Wang J, Frangou C, et al. The Extracellular Niche and Tumor Microenvironment Enhance KRAS Inhibitor Efficacy in Pancreatic Cancer. Cancer Res 2024;84:1115-32. [Crossref] [PubMed]
- Sacher A, LoRusso P, Patel MR, et al. Single-Agent Divarasib (GDC-6036) in Solid Tumors with a KRAS G12C Mutation. N Engl J Med 2023;389:710-21. [Crossref] [PubMed]
- Fakih MG, Salvatore L, Esaki T, et al. Sotorasib plus Panitumumab in Refractory Colorectal Cancer with Mutated KRAS G12C. N Engl J Med 2023;389:2125-39. [Crossref] [PubMed]
- Sarcar B, Tassielli A, Wang L, et al. Abstract B032: KRAS copy number variation and mutant allele fractions predict in vitro response of PDX-derived human pancreatic cancer cell lines to KRASG12D inhibitor MRTX1133. Mol Cancer Res 2023;21:B032. [Crossref]
- Chen Z, Eriksson A, Lee B, et al. Abstract 3340: RMC-5127, a first-in-class, orally bioavailable mutant-selective RASG12V(ON) inhibitor is central nervous system (CNS)-penetrant and demonstrates anti-tumor activity in a preclinical intracranial xenograft model. Cancer Res 2024;84:3340. [Crossref]
- Kirschner T, Müller MP, Rauh D. Targeting KRAS Diversity: Covalent Modulation of G12X and Beyond in Cancer Therapy. J Med Chem 2024;67:6044-51. [Crossref] [PubMed]
- Kim D, Herdeis L, Rudolph D, et al. Pan-KRAS inhibitor disables oncogenic signalling and tumour growth. Nature 2023;619:160-6. [Crossref] [PubMed]
- Burge RA, Hobbs GA. Not all RAS mutations are equal: A detailed review of the functional diversity of RAS hot spot mutations. Adv Cancer Res 2022;153:29-61. [Crossref] [PubMed]


