
Proteome-wide Mendelian randomization and single-cell sequencing analysis identify the association between plasma proteins and gastric cancer
Highlight box
Key findings
• Mendelian randomization (MR) analysis identified 12 plasma proteins with potential causal associations with gastric cancer, enriching the pool of candidate biomarkers for this malignancy.
• Single-cell RNA sequencing analysis revealed distinct expression patterns of these proteins across different stages and cell types of gastric cancer, providing insights into the molecular heterogeneity of the disease.
What is known and what is new?
• Previous studies have highlighted the elevation of certain plasma proteins in patients with gastric cancer, suggesting their potential role in disease pathogenesis.
• This study advances the field by employing a proteome-wide approach to systematically identify plasma proteins with a causal relationship to gastric cancer. The combination of MR and single-cell sequencing analysis in this study is novel, offering a more precise understanding of the genetic architecture of gastric cancer.
What is the implication, and what should change now?
• The identification of novel plasma proteins associated with gastric cancer implicates potential new pathways for disease progression and therapy. These findings advocate for a shift in research focus towards validating and exploring the therapeutic potential of the identified proteins.
Introduction
Background
Gastric cancer is the fifth most common cancer globally and the third leading cause of cancer death (1), resulting in over 784,000 deaths annually (2). The low rate of early diagnosis means that most patients are already in an advanced stage by the time they are diagnosed (3). For these advanced-stage gastric cancer patients, targeted therapy has emerged as a crucial treatment modality. Currently, the main targeted therapies for gastric cancer include anti-human epidermal growth factor receptor 2 (HER2), anti-angiogenesis, and anti-programmed death 1 (PD-1) treatments (4). However, a significant challenge in targeted therapy is the development of drug resistance, which limits the long-term efficacy of these treatments and underscores the need for continuous exploration of new therapeutic targets (5).
Rationale and knowledge gap
Plasma proteins, originating from the active secretion or leakage of tissue cells, allow plasma proteomics to reflect the metabolic activities and genetic characteristics of tissue cells (6). Compared to other approaches, plasma proteomics enables a minimally invasive method for sample collection, which is less burdensome for patients and can be more easily repeated over time to monitor disease progression or response to therapy. Additionally, plasma proteomics can provide insights into systemic disease processes and the overall physiological state, as it captures proteins released into circulation from various tissues, potentially identifying biomarkers that reflect broader pathological changes not confined to a single tissue type.
Previous observational studies have reported several plasma proteins, such as EPHA4 (7) and RUNX3 (8), as elevated in patients with gastric cancer. However, the limitations of these observational studies, including small sample sizes and the influence of confounding factors, have left the causal relationship between these identified proteins and gastric cancer uncertain. Conducting Mendelian randomization (MR) studies with plasma proteome data facilitate a deeper investigation into the genetic factors of diseases (9). In this study, we used protein quantitative trait loci (pQTLs) as instrumental variables (IVs) to infer causal relationships between protein levels and gastric cancer risk. pQTLs are genetic variants that are associated with the levels of specific proteins in the plasma. Since genetic variants are randomly assorted during meiosis and fixed at conception, they are not influenced by confounding factors or reverse causation that typically bias observational studies (10).
Objective
First, we conducted MR analyses using pQTL data for 4,907 proteins from deCODE database (https://www.decode.com) and genome-wide association study (GWAS) data for gastric cancer from the IEU Open GWAS database (https://gwas.mrcieu.ac.uk). Additionally, we utilized single-cell sequencing data from the Gene Expression Omnibus (GEO) database to examine the distribution of potential genetic targets in different cell types and stages of gastric cancer. This study aims to contribute to the understanding of the genetic basis of gastric cancer and to aid in the discovery of novel genetic targets for therapy. We present this article in accordance with the MR-STROBE reporting checklist (11,12) (available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-200/rc).
Methods
MR
Source of GWAS data for gastric cancer and plasma proteome analysis
GWAS data for gastric cancer were downloaded from the IEU GWAS database, identified by the dataset code ebi-a-GCST90018849. This dataset originates from a GWAS study published by Sakaue et al. (13) in 2021. Specifically, the GWAS dataset for gastric cancer encompasses 476,116 individuals, including 475,087 controls and 1,029 cases, with a total of 24,188,662 SNPs, focusing on a European population. The plasma proteomics data were sourced from the study conducted by Ferkingstad et al. (14), which described genome-wide association studies (GWAS) of plasma protein levels measured using 4,907 aptamers across a cohort of 35,559 Icelanders. Given that the original studies for the exposure and outcome data used in this MR study were conducted in different countries, it can be considered that there is no overlap in the populations. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Selection of IVs
The selection of IVs adhered to three widely accepted criteria (15): first, the IVs must demonstrate associations with the exposure, specifically the expression of plasma proteins in this study. Second, the IVs should have no links to potential confounding variables. Third, the IVs are expected to influence the outcome exclusively through known risk factors, without alternative pathways. In MR studies, single nucleotide polymorphisms (SNPs) are commonly employed as IVs. SNPs, which denote single base pair variations in the DNA sequence among individuals, offer several advantages: they are stable throughout an individual’s lifetime, are assorted randomly during the formation of gametes, and can be readily identified (16). These characteristics render SNPs ideal as IVs. In this study, the SNPs employed were mixed pQTLs, including both cis-pQTLs and trans-pQTLs.
To identify SNPs that met the established criteria, we implemented a significance threshold of P <5e-8. To address the issue of linkage disequilibrium (LD) and ensure the independence of SNPs, we utilized a clumping algorithm with parameters set to r2=0.001 and a distance of 10,000 kilobases (kb) by the “TwoSampleMR” R package. To address the concern of bias from weak IVs, the F-statistic for each SNP was calculated, and only those with an F-statistic greater than 10 were retained. Furthermore, in instances where SNPs for the exposure were absent in the outcome datasets, we opted for proxy variants in strong linkage disequilibrium (r2>0.8) with the initial SNPs.
MR analysis
We performed MR analyses for each of the 4,907 plasma proteins to investigate their associations with gastric cancer. Statistical analyses were conducted using R software 4.3.2, with MR analyses performed via the “TwoSampleMR” package (17). The inverse variance weighted (IVW) method served as the primary approach for assessing the causal link between plasma proteins and gastric cancer. This method involves a weighted regression of SNP-outcome associations against SNP-exposure associations, where the intercept is presumed to be zero (18). For instances where only a single compliant SNP was available for a specific protein, the Wald ratio method was applied. To minimize the occurrence of false positives, we employed the false discovery rate (FDR) method for P value adjustment. Plasma proteins with adjusted P values less than 0.05 were defined as significantly positive.
To ensure the validity and robustness of our findings, sensitivity analyses were undertaken, including heterogeneity analysis and pleiotropy analysis. The pleiotropy analysis primarily investigates the presence of horizontal pleiotropy among multiple IVs, using the intercept from the MR-Egger method to identify directional pleiotropy, signified by a significant deviation of the intercept from zero (19,20). Cochran’s Q value, utilized in heterogeneity analysis, measures the variance among IVs, indicating substantial variability when the value is high (21).
Finally, SMR software (22,23) was utilized to conduct summary-data-based MR (SMR) analyses on proteins positive in the MR analysis. This approach tests whether the effects of SNPs on phenotypes are mediated through gene expression. P values were adjusted using FDR correction. The details of MR in this study were not pre-registered in any website.
Single-cell RNA sequencing (scRNA-seq) analysis
Source of scRNA-seq
We employed the Seurat R package (24) for the scRNA-seq analysis process. scRNA-seq data for gastric cancer patients and control individuals were sourced from a study by Kumar et al. (25), representing one of the largest single-cell cohorts for gastric cancer to date. The data have been made publicly available in the GEO database under accession number GSE183904. This dataset initially comprised 40 samples; however, four samples from peritoneal tissues were excluded from our analysis. The remaining 36 gastric tissue samples included both normal gastric tissues and gastric cancer tissues, with the cancer samples spanning stages I through IV.
Quality control
Quality control at the cellular level was conducted using the following criteria: firstly, cells with total unique molecular identifier (UMI) counts below 1,000 were excluded. Secondly, cells expressing fewer than 500 genes were removed. Thirdly, cells exhibiting more than 20% of their total expression from mitochondrial genes were discarded. Fourthly, cells with erythrocyte genes expression exceeding 1% were also eliminated. These thresholds are commonly used in single-cell RNA sequencing studies to balance the inclusion of high-quality cells while minimizing the inclusion of poor-quality or contaminated cells.
Data integration and cell type annotation
For each sample, gene expression levels were quantified as the fraction of total gene expression, subsequently multiplied by 10,000. To facilitate normalization, each value was incremented by 1 (to preclude the logarithm of zero) before conversion into natural logarithms. From the normalized expression matrix, the top 2,000 highly variable genes were selected, then centered and scaled to ensure uniformity in data interpretation. To mitigate batch effects, the Harmony R package (26) was employed, utilizing the principal components analysis (PCA) to adjust based on the leading 15 PCA components. Clustering was conducted utilizing the integrated joint embedding generated by Harmony, following the construction of a shared nearest-neighbor graph via the Louvain algorithm, implemented through the Seurat FindClusters function. The resultant clusters were then visualized on a two-dimensional map using the t-distributed stochastic neighbor embedding (t-SNE) method. For cell type annotation, the SingleR R package (27) was employed, facilitating identification of cell populations within the dataset. Additionally, the receiver operating characteristic (ROC) method was applied to discover marker genes for each cell type. Subsequently, focusing exclusively on tumor tissues, the ROC method was employed once more to identify marker genes for each cell type to determine which genes were enriched in each cell type in tumor cells. We depicted the distribution of encoding genes for plasma proteins identified via MR method in tumor tissues by feature plots.
Differential expression analysis
We designated normal tissues as the control group and tumor tissues as the experimental group, calculating differentially expressed genes (DEGs) for various cell types between the control and experimental groups. The Wilcoxon test was used, and genes with an adjusted P value (after FDR correction) less than 0.05 and an absolute log2 fold change (log2FC) greater than 0.5 were identified as DEGs. DEGs with an adjusted P value less than 0.01 were classified as highly significant; otherwise, they were considered as lowly significant. For the plasma proteins related with gastric cancer identified by MR analyses, we utilized violin plots and box plots to illustrate the expression differences of their encoding genes across various stages of gastric cancer tissues, regardless of whether these genes were classified as DEGs.
Results
MR analysis identified 12 plasma proteins associated with GC
We performed MR analyses for 4,907 plasma proteins and adjusted the P values using FDR correction. Detailed associations between SNPs, exposures, and outcomes, as well as the MR results for each of the 4,907 proteins, are presented in table available at https://cdn.amegroups.cn/static/public/jgo-24-200-1.pdf. The analysis identified 12 plasma proteins significantly associated with gastric cancer. The associations are visualized in a Manhattan plot (Figure 1). Among these, C1GALT1C1 showed a positive correlation with gastric cancer, while CDH1, CHST15, EPHA4, EPHB4, IGF1R, KDR, LIFR, motilin (MLN), OLFM1, SEMA6B, and THSD1 (thrombospondin type 1 domain containing 1) were negatively associated. No evidence of heterogeneity, pleiotropy, or reverse causation was detected for these proteins, ensuring the robustness of our findings. Subsequent SMR analyses confirmed significant associations for MLN and THSD1, with adjusted P values below 0.05 (Table 1).
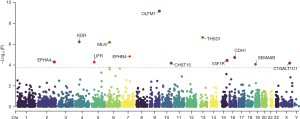
Table 1
| Protein | P for MR | FDR for MR | P for heterogeneity test | P for pleiotropy test | P for SMR | FDR for SMR |
|---|---|---|---|---|---|---|
| C1GALT1C1 | 6.60E−05 | 0.03 | 0.40 | 0.25 | – | – |
| CDH1 | 1.92E−05 | 0.01 | 0.37 | 0.94 | – | – |
| CHST15 | 6.75E−05 | 0.03 | 0.43 | 0.15 | 0.80 | 0.93 |
| EPHA4 | 5.13E−05 | 0.02 | 0.66 | 0.06 | 0.93 | 0.93 |
| EPHB4 | 1.53E−05 | 0.01 | 0.96 | 0.34 | 0.61 | 0.87 |
| IGF1R | 3.66E−05 | 0.02 | 0.68 | 0.22 | 0.53 | 0.87 |
| KDR | 5.97E−07 | 0.0007 | 0.97 | 0.35 | 0.36 | 0.71 |
| LIFR | 5.38E−05 | 0.02 | 0.65 | 0.13 | 0.85 | 0.93 |
| MLN | 6.49E−07 | 0.0007 | 0.14 | 0.30 | 3.72E−06 | 3.72E−05 |
| OLFM1 | 6.82E−10 | 2.81E−06 | 0.37 | 0.56 | 0.14 | 0.48 |
| SEMA6B | 8.64E−05 | 0.03 | 0.69 | 0.76 | 0.28 | 0.69 |
| THSD1 | 2.25E−07 | 0.0005 | 0.53 | 0.13 | 0.00 | 0.01 |
The table shows that all 12 proteins were positively identified in the MR analysis, with no evidence of heterogeneity or pleiotropy. THSD1 and MLN passed the SMR test, and C1GALT1C1 and CDH1 lacked relevant data for SMR analysis. MR, Mendelian randomization; SMR, summary-based Mendelian randomization; FDR, false discovery rate.
scRNA-seq analysis of gastric cancer tissues
scRNA-seq analysis was performed on 145,805 cells from 36 gastric tissue samples, including 10 samples of normal tissue, 3 samples of stage I gastric cancer, 6 samples of stage II gastric cancer, 14 samples of stage III gastric cancer, and 3 samples of stage IV gastric cancer.
Cells were annotated into nine types using the SingleR package: T cells, B cells, endothelial cells, epithelial cells, monocytes, macrophages, neutrophils, smooth muscle cells, and natural killer (NK) cells. The t-SNE plot (Figure 2A) illustrates the clustering of these cell types.
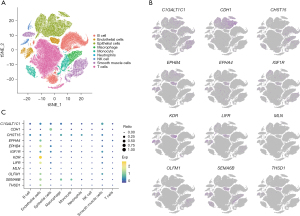
Distribution of identified genes in tumor tissues
The expression analysis of genes corresponding to the 12 identified plasma proteins revealed specific enrichment patterns. EPHB4, KDR, and SEMA6B were enriched in endothelial cells, CDH1 in epithelial cells, and C1GALT1C1 in smooth muscle cells within tumor tissues (Figure 2B,2C). These patterns highlight the potential cell-type-specific roles of these genes in gastric cancer. The remaining genes did not exhibit specific cell-type enrichment but were expressed across various cell types within the tumor tissues.
The enrichment of proteins in certain cell types has important biological implications. For instance, KDR is predominantly expressed in endothelial cells and plays a critical role in angiogenesis, the process of new blood vessel formation. This is particularly relevant in the context of cancer, as tumors require the formation of new blood vessels to grow and metastasize. Similarly, the enrichment of CDH1 in epithelial cells highlights its role in cell-cell adhesion and maintaining epithelial integrity, which is often disrupted in cancer.
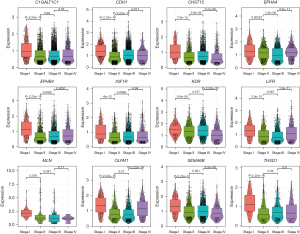
Differential expression across cancer stages
Differential expression analysis showed that KDR and LIFR were significantly downregulated in gastric cancer tissues compared to normal tissues, with adjusted P values less than 0.01. The other ten genes did not show significant differences between normal and cancer tissues. However, all 12 genes exhibited stage-specific expression patterns. For instance, THSD1, CHST15, and SEMA6B had higher expression in stage I compared to later stages, while OLFM1 was more expressed in stages I and IV compared to stages II and III (Figure 3). These findings suggest dynamic changes in gene expression across different stages of gastric cancer progression.
Discussion
In this study, we utilized plasma proteomics data and gastric cancer GWAS data to perform MR analyses, identifying 12 plasma proteins with potential causal associations with gastric cancer. These proteins exhibited neither heterogeneity nor pleiotropy. Subsequently, we conducted single-cell sequencing analyses on 36 gastric tissue samples from GEO database to investigate the distribution of the 12 genes across different cell types and stages of tissue.
Among the 12 proteins and their corresponding genes identified in our study, 7 have been previously reported to be potentially associated with gastric cancer, including CDH1 (28,29), EPHA4 (30,31), EPHB4 (32,33), KDR (34), LIFR (35), IGF1R (36), and SEMA6B (37). Notably, KDR (also known as VEGFR2) is widely recognized for its overexpression in various malignancies (38). There are already inhibitors targeting KDR, such as Ramucirumab, available on the market for targeted therapy in gastric cancer (1). However, our MR analysis inversely found that individuals with lower plasma levels of KDR have a reduced risk of gastric cancer, and our scRNA-seq analysis also indicated a decrease in KDR expression in gastric cancer tissues, which seems to deviate from the established understanding. A potential explanation for this finding could be that although there are no reliable reports on the dual role of the KDR gene in tumor development and progression, many genes and pathways, such as the Keap1–Nrf2 pathway (39) and the MKK3 gene (40), have been reported as having the capability to act both as tumor suppressors and as promoters, depending on the stage of tumor development. This highlights the complexity of targeting specific pathways in cancer therapy. We hypothesize that KDR may exhibit similar characteristics. Another explanation for the discrepancy could be that the gene regulatory mechanisms affecting KDR expression and activity, such as post-translational modifications and interactions with other signaling pathways, might differ between the bloodstream and the tumor microenvironment, leading to differing impacts on cancer progression. In addition, the inherent heterogeneity of gastric cancer, with different genetic and molecular profiles across patients and even within different tumor regions in the same patient, could contribute to the discrepancy. We recognize the need for future studies to better understand the specific role of KDR in gastric cancer.
For the remaining five proteins or genes (including C1GALT1C1, CHST15, THSD1, MLN and OLFM1), there are no reports of their potential to cause gastric cancer to date. Notably, both MLN and THSD1 passed the SMR analysis, suggesting a higher likelihood of a causal relationship with gastric cancer. In scRNA-seq analyses, the expression of THSD1 was significantly lower in stages II, III, and IV of gastric cancer compared to stage I; the expression of MLN was significantly lower in stages III and IV compared to stages I and II. These findings indicate the potential of MLN and THSD1 as novel biomarkers or therapeutic targets for gastric cancer.
THSD1 is a member of the thrombospondin family, associated with angiogenesis. It regulates the formation of blood vessels through the adhesion and migration of endothelial cells (41). Although there have been no previous reports linking THSD1 to gastric cancer specifically, research has reported its downregulation in esophageal cancer tissues, and the absence of THSD1 has been associated with tumor metastasis in esophageal cancer (42). It found that THSD1 downregulation in esophageal cancer was caused by mechanisms such as loss of heterozygosity and promoter hypermethylation, which affected tumor angiogenesis and cell adhesion, suggesting its potential role in cancer progression. We speculate that its involvement in gastric cancer development might be through similar mechanisms.
MLN plays a crucial role in gastrointestinal function, influencing gastric motility through the stimulation of intermittent contractions in the antrum and duodenum (43). A reduction in MLN expression can lead to insufficient gastrointestinal motility (44). Previous research suggests that prolonged retention of gastric contents may increase the duration of gastric mucosal exposure to stomach acid and pepsin, which may lead to damage to gastric mucosa (45), playing a role in the development of gastric cancer (46). Therefore, we hypothesize that the downregulation of MLN may promote the occurrence of gastric cancer by affecting gastric motility which leads to chronic damage to mucosa.
The strength of this study lies in its pioneering use of MR to evaluate the potential causal associations between 4,907 plasma proteins and gastric cancer, complemented by sensitivity and SMR analyses to enhance the robustness of the results. Furthermore, by utilizing the largest scale of scRNA-seq data for gastric cancer available to date, we have characterized the distribution of the 12 genes across different samples, stages, and cell types, corroborating the findings from the MR analysis. This study contributes to the identification of new biomarkers and therapeutic targets for gastric cancer.
Inevitably, this study has several limitations: firstly, the GWAS and scRNA-seq data utilized were derived from European samples, warranting caution when generalizing the study’s findings to other populations. Secondly, the scRNA-seq data mainly reflect the transcriptomic level, which may not fully indicate the expression levels of proteins. Lastly, our hypotheses regarding the mechanisms underlying the associations between KDR, MLN, THSD1, and gastric cancer are based on previous research and our deductions. However, these hypotheses require further investigation by other researchers.
Conclusions
This study identified 12 plasma proteins with potential causal associations with gastric cancer, among which MLN and THSD1 passed the SMR analysis. These proteins can serve as candidate biomarkers or therapeutic targets for gastric cancer and contribute to the etiological and genetic research of the disease. We encourage further investigation into our findings and hypotheses by researchers.
Acknowledgments
The authors would like to thank the contributors to the public databases.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MR-STROBE reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-200/rc
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-200/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-200/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Smyth EC, Nilsson M, Grabsch HI, et al. Gastric cancer. Lancet 2020;396:635-48. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Song Z, Wu Y, Yang J, et al. Progress in the treatment of advanced gastric cancer. Tumour Biol 2017;39:1010428317714626. [Crossref] [PubMed]
- Patel TH, Cecchini M. Targeted Therapies in Advanced Gastric Cancer. Curr Treat Options Oncol 2020;21:70. [Crossref] [PubMed]
- Wadhwa R, Song S, Lee JS, et al. Gastric cancer-molecular and clinical dimensions. Nat Rev Clin Oncol 2013;10:643-55. [Crossref] [PubMed]
- Suhre K, McCarthy MI, Schwenk JM. Genetics meets proteomics: perspectives for large population-based studies. Nat Rev Genet 2021;22:19-37. [Crossref] [PubMed]
- Wu D, Zhang P, Ma J, et al. Serum biomarker panels for the diagnosis of gastric cancer. Cancer Med 2019;8:1576-83. [Crossref] [PubMed]
- Zou XP, Zhang B, Zhang XQ, et al. Promoter hypermethylation of multiple genes in early gastric adenocarcinoma and precancerous lesions. Hum Pathol 2009;40:1534-42. [Crossref] [PubMed]
- Pietzner M, Wheeler E, Carrasco-Zanini J, et al. Mapping the proteo-genomic convergence of human diseases. Science 2021;374:eabj1541.
- Zheng J, Haberland V, Baird D, et al. Phenome-wide Mendelian randomization mapping the influence of the plasma proteome on complex diseases. Nat Genet 2020;52:1122-31. [Crossref] [PubMed]
- Skrivankova VW, Richmond RC, Woolf BAR, et al. Strengthening the Reporting of Observational Studies in Epidemiology Using Mendelian Randomization: The STROBE-MR Statement. JAMA 2021;326:1614-21. [Crossref] [PubMed]
- Skrivankova VW, Richmond RC, Woolf BAR, et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomisation (STROBE-MR): explanation and elaboration. BMJ 2021;375: [Crossref] [PubMed]
- Sakaue S, Kanai M, Tanigawa Y, et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat Genet 2021;53:1415-24. [Crossref] [PubMed]
- Ferkingstad E, Sulem P, Atlason BA, et al. Large-scale integration of the plasma proteome with genetics and disease. Nat Genet 2021;53:1712-21. [Crossref] [PubMed]
- Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ 2018;362:k601. [Crossref] [PubMed]
- Hwang LD, Lawlor DA, Freathy RM, et al. Using a two-sample Mendelian randomization design to investigate a possible causal effect of maternal lipid concentrations on offspring birth weight. Int J Epidemiol 2019;48:1457-67. [Crossref] [PubMed]
- Hemani G, Zheng J, Elsworth B, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018;7:e34408. [Crossref] [PubMed]
- Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol 2013;37:658-65. [Crossref] [PubMed]
- Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 2015;44:512-25. [Crossref] [PubMed]
- Bowden J, Del Greco M F, Minelli C, et al. Improving the accuracy of two-sample summary-data Mendelian randomization: moving beyond the NOME assumption. Int J Epidemiol 2019;48:728-42. [Crossref] [PubMed]
- Verbanck M, Chen CY, Neale B, et al. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet 2018;50:693-8. [Crossref] [PubMed]
- Wu Y, Zeng J, Zhang F, et al. Integrative analysis of omics summary data reveals putative mechanisms underlying complex traits. Nat Commun 2018;9:918. [Crossref] [PubMed]
- Zhu Z, Zhang F, Hu H, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet 2016;48:481-7. [Crossref] [PubMed]
- Hao Y, Stuart T, Kowalski MH, et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat Biotechnol 2024;42:293-304. [Crossref] [PubMed]
- Kumar V, Ramnarayanan K, Sundar R, et al. Single-Cell Atlas of Lineage States, Tumor Microenvironment, and Subtype-Specific Expression Programs in Gastric Cancer. Cancer Discov 2022;12:670-91. [Crossref] [PubMed]
- Korsunsky I, Millard N, Fan J, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 2019;16:1289-96. [Crossref] [PubMed]
- Aran D, Looney AP, Liu L, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol 2019;20:163-72. [Crossref] [PubMed]
- Hansford S, Kaurah P, Li-Chang H, et al. Hereditary Diffuse Gastric Cancer Syndrome: CDH1 Mutations and Beyond. JAMA Oncol 2015;1:23-32. [Crossref] [PubMed]
- Gamble LA, Heller T, Davis JL. Hereditary Diffuse Gastric Cancer Syndrome and the Role of CDH1: A Review. JAMA Surg 2021;156:387-92. [Crossref] [PubMed]
- Miyazaki K, Inokuchi M, Takagi Y, et al. EphA4 is a prognostic factor in gastric cancer. BMC Clin Pathol 2013;13:19. [Crossref] [PubMed]
- Chiang CW, Lin YS, Chang FL, et al. Single-chain fragment antibody disrupting the EphA4 function as a therapeutic drug for gastric cancer. Biochem Biophys Res Commun 2023;680:161-70. [Crossref] [PubMed]
- Yin J, Cui Y, Li L, et al. Overexpression of EPHB4 Is Associated with Poor Survival of Patients with Gastric Cancer. Anticancer Res 2017;37:4489-97. [PubMed]
- Liersch-Löhn B, Slavova N, Buhr HJ, et al. Differential protein expression and oncogenic gene network link tyrosine kinase ephrin B4 receptor to aggressive gastric and gastroesophageal junction cancers. Int J Cancer 2016;138:1220-31. [Crossref] [PubMed]
- Li T, Yu J, Luo X, et al. VEGFR-2 as a novel predictor of survival in gastric cancer: A systematic review and meta-analysis . Pathol Res Pract 2018;214:560-4. [Crossref] [PubMed]
- Zhao J, Li X, Fu L, et al. lncRNA LIFR AS1 inhibits gastric carcinoma cell proliferation, migration and invasion by sponging miR 4698. Mol Med Rep 2021;23:153. [Crossref] [PubMed]
- Oh SC, Sohn BH, Cheong JH, et al. Clinical and genomic landscape of gastric cancer with a mesenchymal phenotype. Nat Commun 2018;9:1777. [Crossref] [PubMed]
- Ge C, Li Q, Wang L, et al. The role of axon guidance factor semaphorin 6B in the invasion and metastasis of gastric cancer. J Int Med Res 2013;41:284-92. [Crossref] [PubMed]
- Itatani Y, Kawada K, Yamamoto T, et al. Resistance to Anti-Angiogenic Therapy in Cancer-Alterations to Anti-VEGF Pathway. Int J Mol Sci 2018;19:1232. [Crossref] [PubMed]
- Tong YH, Zhang B, Fan Y, et al. Keap1-Nrf2 pathway: A promising target towards lung cancer prevention and therapeutics. Chronic Dis Transl Med 2015;1:175-86. [PubMed]
- Piastra V, Pranteda A, Bossi G. Dissection of the MKK3 Functions in Human Cancer: A Double-Edged Sword? Cancers (Basel) 2022;14:483. [Crossref] [PubMed]
- Haasdijk RA, Den Dekker WK, Cheng C, et al. THSD1 preserves vascular integrity and protects against intraplaque haemorrhaging in ApoE-/- mice. Cardiovasc Res 2016;110:129-39. [Crossref] [PubMed]
- Ko JM, Chan PL, Yau WL, et al. Monochromosome transfer and microarray analysis identify a critical tumor-suppressive region mapping to chromosome 13q14 and THSD1 in esophageal carcinoma. Mol Cancer Res 2008;6:592-603. [Crossref] [PubMed]
- Poitras P, Peeters TL. Motilin. Curr Opin Endocrinol Diabetes Obes 2008;15:54-7. [Crossref] [PubMed]
- Feighner SD, Tan CP, McKee KK, et al. Receptor for motilin identified in the human gastrointestinal system. Science 1999;284:2184-8. [Crossref] [PubMed]
- Park MI, Camilleri M. Gastroparesis: clinical update. Am J Gastroenterol 2006;101:1129-39. [Crossref] [PubMed]
- Meyer AR, Goldenring JR. Injury, repair, inflammation and metaplasia in the stomach. J Physiol 2018;596:3861-7. [Crossref] [PubMed]


