
The unique role of cuproptosis in the prognosis and treatment of rectum adenocarcinoma
Highlight box
Key findings
• A novel cuproptosis-related gene (CRG) signature was established that can predict prognosis in rectum adenocarcinoma (READ).
• Lipoic acid synthetase (LIAS) emerged as a favorable prognostic marker and a potential therapeutic target for READ.
What is known, and what is new?
• READ is a distinct digestive cancer subtype with unique clinical features.
• This study showed the significant role of CRGs in READ prognosis with a focus on immune infiltration, and the prognostic value of LIAS.
What is the implication, and what should change now?
• Understanding the role of CRGs provides deeper insights into READ prognosis and could improve patient stratification.
• LIAS could serve as a promising therapeutic target, offering new opportunities for tailored treatment strategies in READ.
Introduction
In recent decades, the incidence of rectum adenocarcinoma (READ) has continued to increased, which accounted for more than 1.9 million newly diagnosed cancer cases and 940,000 cancer-related deaths (1,2). The clinical diagnosis of READ primarily relies on colonoscopy; however, reliable assessment systems for evaluating the severity and prognosis of READ remain inadequate.
Cuproptosis, a newly identified programmed cell death whose mechanism is not apoptotic (3). This process is mediated by the accumulation of copper ions, which disrupt mitochondrial respiration and the tricarboxylic acid cycle. Elevated serum copper levels have been observed in various types of cancer, including stomach cancer and gallbladder carcinoma (4,5). Functionally, copper ions are the activators of receptor tyrosine kinase-related signaling pathways, which directly or indirectly promote tumor growth and the establishment of the tumor microenvironment (TME) (6). These insights have led to the exploration of novel therapeutic approaches that leverage copper-induced cell death in tumors (7,8). The study of the relationship between copper ions and tumors has gone through a series of processes, including copper overdose-induced cell death, copper ion drug-induced tumor death, high concentrations of copper ions being detected in cancerous tissues (as opposed to paraneoplastic tissues), and the discovery of the genes and pathways of copper-associated death (9-12). In March 2022, Peter Tsvetkov and Todd R. Golub of the Broad Institute of MIT and Harvard University demonstrated that cuproptosis is a novel mode of cell death that is distinct from iron death and apoptosis, in that it relies on mitochondrial respiration and induces lipoylated proteins and iron-sulfur cluster protein loss through the direct binding of copper to thioctic components of the tricarboxylic acid cycle, leading to toxic protein stress. The results of this study were also published in the journal Science (13). Tsvetkov et al. describe the specific details of cuproptosis as (3): (I) excess copper enters the cell; (II) copper ions bind to thioacylation-modifying proteins, FDX1 being an upstream regulatory molecule that regulates the thioacylation pathway; and (III) the four thioacylation-modifying proteins known to date in mammals, DBT (branched-chain α-ketoacid dehydrogenase complex component), GCSH (glycine cleavage system protein H), DLST (α-ketoglutarate dehydrogenase complex component), and DLAT (pyruvate dehydrogenase complex component), all of which are core components of the cellular complex of important metabolic enzymes that regulate carbon entry into the tricarboxylic acid cycle; (IV) oligomerization of lipoic acid-modified proteins bound to copper ions; (V) great impact on the tricarboxylic acid cycling cycle; (VI) reduction in the level of Fe-S cluster proteins (branched pathway); and (VII) Cu2+ ion binds FDX1 and changes to the more toxic Cu+; (VIII) the above processes ultimately lead to cell death. This death pathway cannot be alleviated by apoptotic caspase inhibitors (Z-VAD-FMK and Boc-D-FMK), iron death inhibitors (ferrostatin-1), necrotic apoptosis inhibitors (necrostatin-1) and oxidative stress inhibitors (N-acetylcysteine). This study found that cuproptosis has great potential for application in cancer therapy, and that cuproptosis can be specifically induced in tumor cells by applying a copper ionophore to inhibit cancer (14). Cuproptosis-related genes (CRGs) have been demonstrated prognostic and immunotherapeutic value in multiple cancer types. CRG-based molecular signatures have been used to predict patient prognosis in glioma (15), cholangiocarcinoma (16), and renal cancer (17). Despite growing evidence supporting clinical importance of CRGs in various types of tumors (18,19), their specific role in READ remains unclear. This study aims to investigate the prognostic significance of CRGs in READ, and their association with TME.
Using the consensus clustering, we categorized READ samples into three distinct subtypes. By examining the overlap of differentially expressed genes (DEGs) among these clusters, we developed the cuproptosis-related score (CRS) as an effective means for assessing the prognosis of READ patients. Further comparisons were made between high- and low-CRS groups to assess the differences in the immune status and mutation landscape. Lipoic acid synthetase (LIAS) was identified as a favorable CRG and was found to be closely correlated with the tumor-related signaling pathways and tumor immune landscape.
This study provides new insights into the role of cuproptosis in READ and highlights LIAS as a potential therapeutic target, offering novel strategies for precision oncology. We present this article in accordance with the TRIPOD and MDAR reporting checklists (available at https://jgo.amegroups.com/article/view/10.21037/jgo-2025-105/rc).
Methods
Data acquisition and processing
The Cancer Genome Atlas-rectum adenocarcinoma (TCGA-READ) dataset provided the necessary data for this study. This dataset included RNA-sequencing data, clinical data, and somatic mutation data. Samples with missed clinical messages were deleted, and the batch effects were eliminated. A total of 10 CRGs summarized in a previous study were identified (3). The Wilcoxon method was used to filter the differentially expressed CRGs. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Identification of cuproptosis clusters
The R package ConsensusClusterPlus was used to perform the unsupervised clustering based on the 10 CRGs. The R package ConsensusClusterPlus was installed via Bioconductor using BiocManager::install(“ConsensusClusterPlus”). The package is developed by Wilkerson MD and Hayes DN and is available at Bioconductor. The determination of the number of clusters was based on the results obtained from the consensus clustering calculation (20). Moreover, the prognosis of each cuproptosis cluster was calculated using the Kaplan-Meier method. The principal component analysis (PCA) was performed using the R package pcaMethods. The R package pcaMethods was installed via Bioconductor using BiocManager::install (“pcaMethods”).
Evaluation of the biological function and immune landscape
Using the R packages Gene Set Variation Analysis (GSVA), clusterProfiler and enrichplot, we performed the enrichment analysis of the DEGs. The R packages GSVA, clusterProfiler and enrichplot were available from Comprehensive R Archive Network (CRAN) (https://cran.r-project.org/). The reference gene set for this analysis was obtained from the Molecular Signatures Database (MSigDB) (21).
Immune cell infiltration was estimated based on multiple algorism and results from the Tracking Tumor Immunophenotype (TIP) website, an online website for the analysis of tumor immunophenotype, which analyses anti-cancer immune status across seven steps (steps 1–7) through the tumor immune cycle (22,23). Moreover, the single sample Gene Set Enrichment Analysis (ssGSEA) method was used based on the gene markers from the research of Charoentong et al. (24). Additionally, the overall condition of the TME was assessed using the ESTIMATE algorithm (25).
Establishment of the prognostic score
To better evaluate the prognosis of each patient, the CRS was established as follows: (I) the overlapping DEGs between each cuproptosis cluster were identified; (II) using a Cox regression model, genes with a prognostic value were selected for further investigation. Based on these selected genes, the PCA method was used, and the CRS was calculated as follows (26): CRS = PC1i + PC2i, where i represents the included CRG.
The performance of the prognostic score was evaluated using univariate and multivariate Cox regression analyses. The univariate analysis identified significant predictors of survival, which were then included in the multivariate model to assess their independent contributions.
Identification of genetic alterations and drug sensitivity analysis
The R package maftools was used to generate waterfall plots of single nucleotide variants in TCGA-READ dataset. The R package maftools was available from CRAN. A further analysis was conducted to examine variations in the expression of the tumor mutational burden (TMB) among the different subgroups. Kaplan-Meier curves were used to assess differences in survival between the mutation and the CRS combination, and to further evaluate the association between the mutation and the CRS. For the analysis of targeted therapeutic drugs (27), the R package pRRophetic was employed to predict the half-maximal inhibitory concentration (IC50) values of lapatinib, nilotinib, parthenolide, and rapamycin, which are commonly investigated for their potential therapeutic effects in READ. The R package pRRophetic was installed via Bioconductor using BiocManager::install (“pRRophetic”).
Nomogram construction
Nomograms predicting the 1-, 3-, and 4-year overall survival (OS) rates of READ patients were constructed using the R packages rgplot, survival, and rms (28). Nomograms predicting the 1-, 3-, and 4-year OS rates of READ patients were constructed using the R packages rms, survival and regplot, all of which are freely available from CRAN at https://cran.r-project.org. Calibration curves for the nomograms were also created using the rms package. These packages are developed and maintained by the R Core Team and other contributors under the GNU General Public License.
Cell culture and assay validation
The HCT116 and RKO cell lines of human colorectal cancer were acquired from GeneChem in Shanghai, China. These cell lines were cultivated in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum at a temperature of 37 ℃ in an atmosphere containing 5% carbon dioxide. The HCT116 cells and RKO cells were transfected with the control plasmid and overexpression plasmid of LIAS. The proliferation activity of the colorectal cancer cells was assessed by Cell Counting Kit 8 (CCK-8) and clone formation assays. The migration ability of the colorectal cancer cells was evaluated by Transwell assay.
Statistical analysis
The statistical analyses were performed using R software (version 4.1.2), a free software environment for statistical computing and graphics developed by the R Core Team. The software is available at https://www.r-project.org. All statistical tests were two-sided. A P value <0.05 or a Spearman correlation coefficient >0.2 was considered statistically significant unless otherwise noted. In abbreviated representation, * represents P≤0.05, ** represents P≤0.01, and *** represents P≤0.001.
Results
Identification of cuproptosis clusters in READ
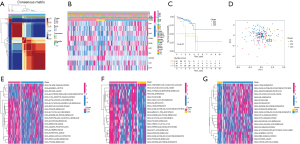
Figure 1A shows the unsupervised consensus clustering results based on the expression of the 10 CRGs. Figure 1B shows the expression signatures of the 10 CRGs, together with the corresponding clinical features. To validate the clustering results, a Kaplan-Meier plot was generated that revealed significant differences in prognosis among the cuproptosis clusters (Figure 1C). The PCA plot showed the fine effectiveness of differentiation (Figure 1D). Cuproptosis type (CT) 1 and CT3 exhibited similar prognostic features. The CT1 cluster was enriched in multiple metabolic pathways, including the glyoxylate and dicarboxylate metabolism pathways, and pyruvate metabolism and arginine and proline metabolism pathways, indicating abrupt metabolic changes in CT1 (Figure 1E). Compared with the CT1 cluster, the CT2 cluster had elevated complement and coagulation cascades (Figure 1F). The CT2 cluster was also enriched in the activated notch signaling pathway, mammalian target of rapamycin (mTOR) signaling pathway, and tumor protein 53 (P53) signaling pathway, which revealed that proliferative activity was elevated in CT2 (Figure 1G). These differences in biological pathways ultimately resulted in the CT2 cluster showing the worst survival.
Establishment of the cuproptosis-related prognostic model
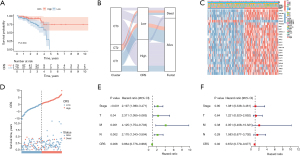
To assess the predictive significance of cuproptosis in READ, we created a PCA model called the CRS using the overlapping genes among the three cuproptosis clusters. The patients were divided into high- and low-CRS groups based on a moderate cut-off value. Figure 2A shows the Kaplan-Meier survival curve, which revealed that patients in the high-CRS group had better OS than those in the low-CRS group. The low-CRS group comprised a larger proportion of CT1 samples and all the CT2 samples, and the survival of this group was significantly worse than that of the high-CRS group (Figure 2B). Figure 2C shows the clinical parameters and expression patterns of the encapsulated genes for each sample, while Figure 2D shows the distribution of the CRS and survival time. Following both the univariate and multivariate Cox analyses, the CRS was found to be an independent protective factor associated with READ prognosis in comparison to other conventional prognostic factors (all P<0.05, hazard ratio <1, Figure 2E,2F). Although the M and N stages are well-recognized prognostic factors in colorectal cancer, they did not reach statistical significance in our multivariate analysis. This suggests that CRS may capture key tumor characteristics, including metabolic and molecular features, which provide complementary or even superior prognostic value compared to traditional clinicopathological factors. Therefore, CRS could serve as a more comprehensive prognostic indicator in READ.
Comparison of the immune landscape between the high- and low-CRS groups
Multiple methods were used to estimate the infiltration or proportion of immune cells (Figure 3A). Specifically, the CRS exhibited a positive correlation with the estimated B cells, macrophages, and T helper cells, but a negative correlation with regulatory T cells (Tregs) (Figure 3B). The low-CRS group exhibited a notable increase in the infiltration of Tregs, myeloid-derived suppressor cells (MDSCs), and macrophages, which were recognized as significant immunosuppressive elements in the TME (Figure 3C). Additionally, the low-CRS group had a significantly higher ImmuneScore and ESTIMATEScore, indicating more immune cells and tumor cell infiltration (Figure 3D). The ssGSEA results showed that the low-CRS group had a higher level of immune checkpoint signatures, and increased cytolytic activity, and human leukocyte antigen molecules compared to the high-CRS group (Figure 3E). Moreover, the results from the tumor infiltrating lymphocyte (TIL) website indicated a higher infiltration of T helper cells in the high-CRS group, and a higher infiltration of Tregs and MDSCs in the low-CRS group (Figure 3F). Together, these results showed that the immune regulatory cells, including the Tregs and MDSCs, were increased in the low-CRS group, which may lead to the anergy of immune effector cells.
CRS subgroups had significant clinical implications and unique mutation landscapes
We also investigated the association between the CRS and the clinical stages of READ. Notably, the CRS was significantly lower in the advanced-stage samples and metastasis samples, and steadily decreased in patients with advanced T and N stages (Figure 4A). Further, the low-CRS group had a significantly greater percentage of patients with advanced READ (all P<0.05, Figure 4B). In terms of patient prognosis, for patients with T3–4, N1–2, and stage III–IV clinical status, the high-CRS group had significantly better OS than the low-CRS group (Figure 4C). To facilitate a clear comparison between early-stage and advanced-stage colorectal cancer, we grouped stage III and IV together, following a common approach. This classification highlights overall prognostic differences between low-risk and high-risk groups, providing a clearer understanding of survival trends. The decrease in the number of patients at risk over time in survival analysis is an expected outcome in long-term follow-up studies due to disease progression, loss to follow-up, or censored cases. Despite this, our follow-up period aligns with similar studies, and the observed trends support our findings. The unexpectedly good prognosis of the high-CRS group in M1 patients is likely due to the small sample size rather than factors such as radical metastasis resection rates or insufficient follow-up. Figure S1A,S1B shows the overall mutation landscape of the high- and low-CRS groups, respectively. The low-CRS group had a reduced TMB compared to the high-CRS group (Figure S1C). Additionally, the PCA results showed that the TMB was positively correlated with the CRS (R=0.26, P=0.0014, Figure S1D). Patients with a higher TMB had better OS than those with a lower TMB (P<0.001, Figure S1E). Together, the CRS and TMB could be used to predict patient prognosis (P<0.001, Figure S1F).
LIAS was the hub gene of CRGs in READ
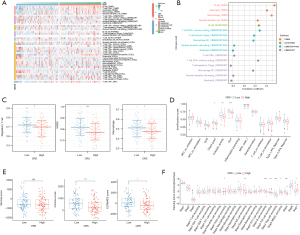
To identify the hub CRGs in READ, we conducted further investigations to determine their predictive significance and association with immune markers. Of the 10 CRGs, seven (CDKN2A, DLAT, DLD, FDX1, GLS, PDHA1, and PDHB) displayed increased expression in tumor tissues. Conversely, LIAS displayed decreased expression (Figure S2A). Moreover, the low-LIAS expression group had significantly worse survival compared to high-LIAS group (Figure S2B-S2K). To evaluate the individual role of LIAS in human cancers, a pancancer expression analysis was conducted. As Figure 5A shows, LIAS was differentially expressed in 10 other cancer types (P<0.05). In half of the 10 types of cancers, LIAS was highly expressed in tumor tissues compared to normal tissues, including bile duct cancer (CHOL), liver cancer (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), and stomach cancer (STAD). In the remaining five cancer types, LIAS was lowly expressed in tumor tissues compared with normal tissues, including breast cancer (BRCA), kidney chromophobe (KICH), kidney clear cell carcinoma (KIRC), thyroid cancer (THCA), esophageal cancer (ESCA) and endometrial cancer (UCES). Further, the immunofluorescent staining of the human cell line showed that LIAS was mainly localized to the nucleoplasm (Figure 5B,5C).
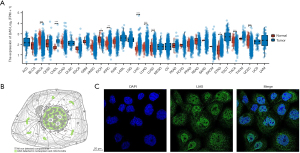
LIAS was involved in a variety of cancers and immune-related signaling pathways
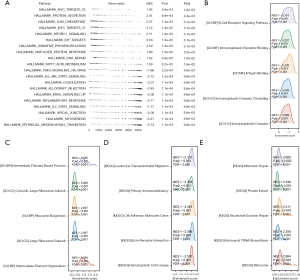
The GSEA method was employed to examine differences in the pathways enriched by the DEGs between the samples with high and low LIAS expression. The findings from the HALLMARK pathway enrichment analysis indicated that the genes regulated by LIAS were linked to the regulation of different processes and functions related to tumors, including increased activity in pathways such as the myelocytomatosis oncogene (myc) signaling pathway, E2F transcription factor (E2F) signaling pathway, and mTOR signaling pathway, and reduced activity in pathways such as the interleukin-6 (IL-6)-Janus kinase (JAK)-signal transducer and activator of transcription 3 (STAT3) signaling pathway and the Kirsten rat sarcoma viral oncogene homolog (Kras) signaling pathway (Figure 6A). In relation to the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) pathways, the decreased expression of LIAS also enhanced immune-associated signaling pathways, including the B cell receptor signaling pathway, immunoglobulin receptor binding, and primary immunodeficiency (Figure 6B,6C). Conversely, the significant overexpression of LIAS primarily enriched the signaling pathways related to the synthesis and transport of amino acids and proteins, such as ribosome biogenesis, aminoacyl transfer RNA biosynthesis, and protein export (Figure 6D,6E).
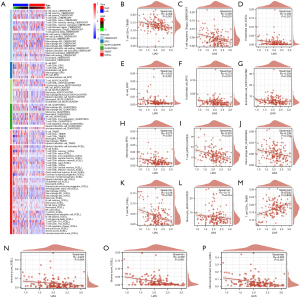
Additionally, we examined the infiltration of immune cells in the high- and low-LIAS expression subtypes using six different algorithms (TIMER, CIBERSORT, MCPCOUNTER, XCELL, QUANTISEQ and EPIC), and compared the results (Figure 7A). In our findings, we observed a negative correlation between LIAS expression and the abundance of various immune cells infiltrating the system, including important immunosuppressive cells like Tregs and M2 macrophages (Figure 7B-7L). Conversely, we observed a positive correlation between LIAS expression and the abundance of infiltrating cluster of differentiation (CD)8+ T cells (Figure 7M). Further, LIAS expression was negatively correlated with the immune score, stromal score, and microenvironment score (Figure 7N-7P). These findings suggest that LIAS could have a distinct function in immune infiltration, and could serve as a predictor for the immunotherapy response in READ.
LIAS was associated with the FBXW7 mutation in READ
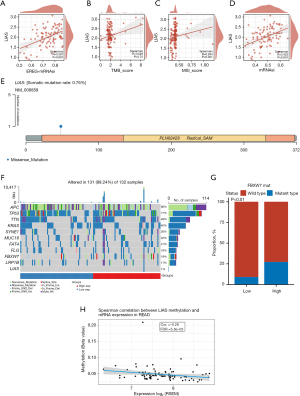
We examined the effect of LIAS on genomic alterations (Figure 8A-8D). We observed a positive correlation between LIAS and epigenetically regulated (EREG)-mRNA stemness index (mRNAsi) (R=0.367, P<0.001, Figure 8A), microsatellite instability-high (MSI) (R=0.264, P=0.001, Figure 8C), and mRNAsi (R=0.422, P<0.001, Figure 8D). Conversely, no significant correlation was observed between the TMB and LIAS (Figure 8C). Additionally, our study revealed the mutation patterns associated with LIAS in READ (Figure 8E-8G). Specifically, the prevalence of FBXW7 mutations was higher in patients with elevated LIAS expression (Figure 8G). In terms of methylation of READ, it can be found that the degree of LIAS methylation is inversely proportional to the degree of expression and is statistically significant (Figure 8H). In terms of the clinical correlations, the patients with FBXW7 mutations exhibited potentially better clinical characteristics than those with the wild-type patients (Figure 9A). The GSEA results showed that the FBXW7-mutation patients displayed increased activity in the E2F target, mTORC1 signaling, oxidative phosphorylation, and G2M checkpoints, but decreased activity in reactive oxygen species, ultraviolet (UV) response, early estrogen response, transforming growth factor-beta signaling, bile acid metabolism, xenobiotic metabolism, and epithelial mesenchymal transition (Figure 9B). In addition, we created a protein-protein interaction (PPI) network using the DEGs between FBXW7-mutation and wild-type patients (Figure 9C).
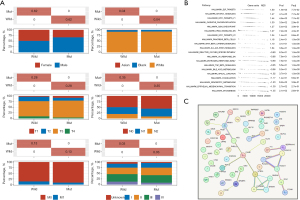
Diagnostic value of LIAS, and drug concentration prediction in READ
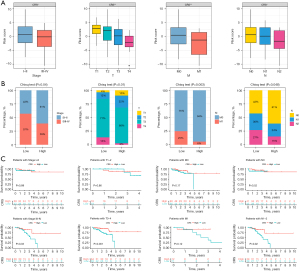
Nomograms serve as robust predictive models in oncology, enabling the estimation of survival probabilities in cancer patients (29). To this end, we first analyzed the differences in LIAS expression in different clinical subgroups. The results showed significant differences in LIAS expression across clinical subgroups. LIAS expression was relatively low in the subgroups of patients with more advanced T, N, and M stages, greater ages, and lymphatic invasion (Figure 10A-10E). Further, we created a nomogram that combined clinical factors and LIAS to estimate the survival chances of patients at 1, 3, and 4 years. The findings indicated that the prognosis prediction, which took into account the level of LIAS expression along with conventional clinical factors such as the pathological T, N, M stages, age, and lymphatic invasion, demonstrated a consistently reliable predictive value (Figure 10F,10G). Moreover, the expression of LIAS could also be used to predict drug sensitivity. As Figure 10H-10K shows, the estimated IC50 scores for lapatinib and parthenolide were lower in the low-LIAS group, implying heightened sensitivity to these agents among individuals with this subtype of READ. Conversely, patients in the high-LIAS group exhibited heightened sensitivity to nilotinib and rapamycin. These results suggest that LIAS expression levels could be used to guide personalized immunotherapy regimens. Patients with low LIAS expression, who exhibit an immunosuppressive environment, may benefit from combination strategies such as immune checkpoint inhibitors (ICIs) combined with mTOR inhibitors (e.g., rapamycin) or metabolic reprogramming therapies targeting oxidative phosphorylation and amino acid metabolism. High-LIAS patients may respond better to ICIs alone or in combination with targeted therapies such as nilotinib.
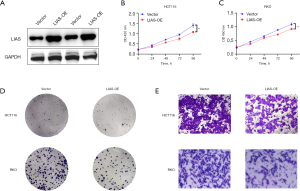
LIAS overexpression inhibited the proliferation and migration abilities of colorectal cancer cells in vitro
To evaluate the tumor-related effect of LIAS in vitro, HCT116 and RKO cells were stably transfected with a plasmid expressing LIAS, while cells transfected with a scrambled vector were used as a negative control. Using the plasmid, LIAS was successfully overexpressed in the two colorectal cancer cell lines (Figure 11A). Subsequently, the effect of LIAS on READ cell proliferation was examined. The CCK-8 assay revealed a significant reduction in the proliferation ability of the HCT116 and RKO cells following the overexpression of LIAS (Figure 11B,11C). These findings were further substantiated by clone assays conducted in both the HCT116 and RKO cells (Figure 11D), indicating that LIAS effectively inhibited the proliferation of READ cells in vitro. Additionally, Transwell assays were performed to assess the effect of LIAS on the migration and invasion abilities of READ cells. The results showed that LIAS inhibited the malignancy of READ cells, as evidenced by the decreased migration and invasion abilities in the HCT116 and RKO cells following LIAS overexpression (Figure 11E).
Discussion
The role of copper-induced cell death, or cuproptosis, has gained increasing attention in recent years. Copper, an essential metal ion for cellular metabolism, becomes cytotoxic when accumulated beyond physiological level, leading to cell death. Dysregulation of copper homeostasis has been implicated in various diseases (30,31). While apoptosis and the autophagy pathway are well established mechanisms of copper-induced cytotoxicity (32), emerging evidence suggests that cuproptosis plays a distinct role in the proliferation, metastasis and angiogenesis of cancer cells (33). Studies have shown that serum copper ion levels are notably elevated in cancer patients compared to healthy individuals, including those with lung cancer, prostate cancer, breast cancer, gallbladder carcinoma, stomach cancer, and thyroid cancer. Additionally, higher copper ion levels have been detected in the gallbladder tissue of patients with gallbladder cancer. Furthermore, in lung cancer patients, elevated serum copper ion concentrations have been correlated with more advanced clinical stages and poorer clinical prognosis (34,35). Although CRGs have been recognized as prognostic marker in multiple cancers, their significance in READ remained unclear. This study aimed to elucidate the role of CRGs in READ prognosis and TME.
Based on the expression patterns of the 10 CRGs and unsupervised clustering, the READ samples were stratified into three clusters exhibiting distinct expression profiles, each associated with significantly varied prognoses. Notably, the CT2 showed the worst survival outcomes, which may be cluster, which was attributed to elevated expression of CDKN2A, a key regulator of cell cycle arrest and senescence. The functional pathway enrichment analysis revealed that CT2 has enriched in tumor promoting pathway, including the notch signaling pathway, the mTOR signaling pathway, and the P53 signaling pathway as well as dysregulated metabolic pathway activity, such as pyruvate metabolism, the citrate cycle, and glutathione metabolism. These findings suggest that cuproptosis heterogeneity contributes to differences in tumor progression and prognosis.
To enhance the prognostic precision, we established the CRS using PCA-based model. CRS effectively stratified READ patients into high- and low-risk groups with higher CRS associated with better OS. Notably, CRS demonstrated a strong inverse correlation with tumor progression, as lower CRS values were observed in patients with advanced clinical stages. Furthermore, when combined with TMB, CRS provided an even more robust prognostic prediction. These findings highlight the potential clinical utility of CRS in risk stratification and personalized treatment planning for READ patients.
Our study also identified LIAS as a key CRG with significant prognostic value. LIAS plays a crucial role in mitochondrial metabolism by regulating protein lipoylation, which is essential for key metabolic enzymes in the TCA cycle. In cuproptosis, a copper-induced form of regulated cell death, LIAS loss confers resistance to copper toxicity, highlighting its role in linking mitochondrial function to cellular stress responses. Disruption of LIAS impairs Fe-S cluster protein integrity and alters metabolic homeostasis, which can influence tumor progression by modulating energy production, oxidative stress, and immune microenvironment interactions. Given its involvement in metabolic reprogramming, LIAS may serve as both a tumor vulnerability and a potential therapeutic target in cancers where cuproptosis-related pathways are dysregulated (36). LIAS expression was notably decreased in the READ tissue compared with the normal tissue, and low LIAS expression was significantly associated with a poor survival, which suggests that LIAS may play a protective role in READ. Cai et al. conducted a pancancer study to explore the role of LIAS, and found that high LIAS predicted better OS and disease-free survival (DFS) in 92 READ samples (37), which is consistent with our results based on 158 READ samples.
LIAS has been implicated in the synthesis of mitochondria-associated metabolic enzymes, and has been found to play a role in energy metabolism and the antioxidant response (38). Over the past decade, research has focused on the effects of LIAS mutations on mitochondrial energy-consuming metabolism, which can lead to complex metabolic disorders (39). However, its potential mechanisms and biological functions in cancer remain insufficiently explored. As our study showed, the LIAS-associated genes in the READ samples were predominantly enriched in the tumor-associated signaling pathways, including the myc, mTOR-related, and IL-6-JAK-STAT pathways. Burr et al. showed that the disruption of LIAS led to the accumulation of 2-oxoglutarate and the subsequent production of L-2-hydroxyglutarate (L-2-HG). In the presence of oxygen, L-2-HG had the ability to stabilize hypoxia inducible factor-1α (HIF-1α) by blocking prolyl hydroxylation, resulting in the initiation of subsequent responses by HIF-1α (40). LIAS deficiency in individuals with impaired lipoic acid synthesis may trigger HIF-1α activation, which may serve as a defensive reaction against metabolic disorders. This phenomenon could potentially account for the reduced LIAS expression observed in certain cancer cells like renal clear cell carcinoma cells, where the involvement of HIF and VHL genes is crucial for tumor development (37).
In addition, in the low-LIAS subgroup, there were more infiltrated Tregs, MDSCs, and macrophages, which were considered the main immunosuppressive cells in the TME. Conversely, the high-LIAS subgroup exhibited an increased presence of immune effector cells such as CD8+ T cells. These findings underscore the potential significance of LIAS in shaping the immune landscape in the TME. Our results also revealed a possible correlation between LIAS expression and the FBXW7 mutation. Interestingly, the FBXW7-mutant colorectal cancer patients had better DFS than the FBXW7 wild-type patients. Additionally, the FBXW7 mutation has been shown to be correlated with shorter DFS in lung squamous cell carcinoma, ovarian plasmacytoma, and bladder urothelial carcinoma (41). Such findings suggest that the FBXW7 mutation plays a special role in colorectal cancer. Our results also provide further evidence of the possible protective role of the FBXW7 mutation in READ.
The atypical manifestation of genes (29) could facilitate cancer diagnosis and prognosis prediction. In this study, the nomogram analysis indicated that LIAS expression combined with classical clinicopathological features had significant diagnostic value for READ patients, and could effectively predict the 1-, 3-, and 4-year survival probabilities of individuals. Interestingly, we also found differences in the predicted concentrations of commonly prescribed drugs among different LIAS expression subgroups, which suggests that LIAS also has value in predicting drug sensitivity.
Finally, to substantiate the biological function of LIAS in READ cells, we conducted LIAS overexpression experiments in two colorectal cancer cell lines (HCT116 and RKO). Our findings revealed that heightened LIAS expression led to a significant reduction in the growth potential of both the HCT116 and RKO cell lines. Additionally, Transwell assays revealed a notable decrease in the migratory ability of the colorectal tumor cells. Consequently, our results suggest that LIAS plays a role in inhibiting the progression of READ, shedding light on its potential biological significance in READ pathogenesis.
This study highlighted the potential prognostic value of CRGs in READ; however, it had several limitations. First, the sample size was relatively small, and further research needs to be conducted to assess the generalizability and reliability of the model. Second, the specific mechanisms of LIAS and its associated pathways in READ require further investigation to fully elucidate their biological functions.
Conclusions
This study highlights the prognostic significance of CRGs in READ, establishing CSR as a novel prognostic tool. Furthermore, LISA emerged as a key tumor suppressor and potential immunotherapy target. By integrating CRS and LIAS-based biomarkers, clinicians may improve risk stratification and optimize personalized treatment. Strategies for READ patients. Future research should focus on validating these findings in larger cohorts and functional in vivo studies to facilitate clinical translation.
Acknowledgments
None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD and MDAR reporting checklists. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-2025-105/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-2025-105/dss
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-2025-105/prf
Funding: This work was supported by grants from
Conflicts of Interests: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-2025-105/coif). The authors have no conflict of interests to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Dekker E, Tanis PJ, Vleugels JLA, et al. Colorectal cancer. Lancet 2019;394:1467-80. [PubMed]
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [PubMed]
- Tsvetkov P, Coy S, Petrova B, et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 2022;375:1254-61. [PubMed]
- Basu S, Singh MK, Singh TB, et al. Heavy and trace metals in carcinoma of the gallbladder. World J Surg 2013;37:2641-6. [PubMed]
- Yaman M, Kaya G, Yekeler H. Distribution of trace metal concentrations in paired cancerous and non-cancerous human stomach tissues. World J Gastroenterol 2007;13:612-8. [PubMed]
- He F, Chang C, Liu B, et al. Copper (II) Ions Activate Ligand-Independent Receptor Tyrosine Kinase (RTK) Signaling Pathway. Biomed Res Int 2019;2019:4158415. [PubMed]
- Xie J, Yang Y, Gao Y, et al. Cuproptosis: mechanisms and links with cancers. Mol Cancer 2023;22:46. [Crossref] [PubMed]
- Wang T, Liu Y, Li Q, et al. Cuproptosis-related gene FDX1 expression correlates with the prognosis and tumor immune microenvironment in clear cell renal cell carcinoma. Front Immunol 2022;13:999823. [Crossref] [PubMed]
- Deng Y, Chi P, Lan P, et al. Modified FOLFOX6 With or Without Radiation Versus Fluorouracil and Leucovorin With Radiation in Neoadjuvant Treatment of Locally Advanced Rectal Cancer: Initial Results of the Chinese FOWARC Multicenter, Open-Label, Randomized Three-Arm Phase III Trial. J Clin Oncol 2016;34:3300-7. [Crossref] [PubMed]
- Loree JM, Wang Y, Syed MA, et al. Clinical and Functional Characterization of Atypical KRAS/NRAS Mutations in Metastatic Colorectal Cancer. Clin Cancer Res 2021;27:4587-98. [Crossref] [PubMed]
- Ratovomanana T, Nicolle R, Cohen R, et al. Prediction of response to immune checkpoint blockade in patients with metastatic colorectal cancer with microsatellite instability. Ann Oncol 2023;34:703-13. [Crossref] [PubMed]
- Wolf AMD, Fontham ETH, Church TR, et al. Colorectal cancer screening for average-risk adults: 2018 guideline update from the American Cancer Society. CA Cancer J Clin 2018;68:250-81. [Crossref] [PubMed]
- Biller LH, Schrag D. Diagnosis and Treatment of Metastatic Colorectal Cancer: A Review. JAMA 2021;325:669-85. [Crossref] [PubMed]
- Patel SG, Karlitz JJ, Yen T, et al. The rising tide of early-onset colorectal cancer: a comprehensive review of epidemiology, clinical features, biology, risk factors, prevention, and early detection. Lancet Gastroenterol Hepatol 2022;7:262-74. [Crossref] [PubMed]
- Wang W, Lu Z, Wang M, et al. The cuproptosis-related signature associated with the tumor environment and prognosis of patients with glioma. Front Immunol 2022;13:998236. [Crossref] [PubMed]
- Chen J, Yu X, Tong H, et al. Establishment and experimental validation of a novel cuproptosis-related gene signature for prognostic implication in cholangiocarcinoma. Front Oncol 2022;12:1054063. [Crossref] [PubMed]
- Sun P, Xu H, Zhu K, et al. The cuproptosis related genes signature predicts the prognosis and correlates with the immune status of clear cell renal cell carcinoma. Front Genet 2022;13:1061382. [Crossref] [PubMed]
- Sheng H, Gu J, Huang Y, et al. Cuproptosis-related signature predicts prognosis and indicates tumor immune infiltration in bladder cancer. Transl Androl Urol 2024;13:2280-93. [Crossref] [PubMed]
- Ma S, Xu M, Zhang J, et al. Analysis and functional validations of multiple cell death patterns for prognosis in prostate cancer. Int Immunopharmacol 2024;143:113216. [Crossref] [PubMed]
- Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 2010;26:1572-3. [PubMed]
- Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [PubMed]
- Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity 2013;39:1-10. [PubMed]
- Xu L, Deng C, Pang B, et al. TIP: A Web Server for Resolving Tumor Immunophenotype Profiling. Cancer Res 2018;78:6575-80. [PubMed]
- Charoentong P, Finotello F, Angelova M, et al. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep 2017;18:248-62. [PubMed]
- Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4:2612. [PubMed]
- Mariathasan S, Turley SJ, Nickles D, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554:544-8. [PubMed]
- Geeleher P, Cox N, Huang RS. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One 2014;9:e107468. [PubMed]
- Zhang Z, Kattan MW. Drawing Nomograms with R: applications to categorical outcome and survival data. Ann Transl Med 2017;5:211. [PubMed]
- Jin CY, Du L, Nuerlan AH, et al. High expression of RRM2 as an independent predictive factor of poor prognosis in patients with lung adenocarcinoma. Aging (Albany NY) 2020;13:3518-35. [PubMed]
- Chen L, Min J, Wang F. Copper homeostasis and cuproptosis in health and disease. Signal Transduct Target Ther 2022;7:378. [PubMed]
- Garza NM, Swaminathan AB, Maremanda KP, et al. Mitochondrial copper in human genetic disorders. Trends Endocrinol Metab 2023;34:21-33. [PubMed]
- Xue Q, Kang R, Klionsky DJ, et al. Copper metabolism in cell death and autophagy. Autophagy 2023;19:2175-95. [PubMed]
- Wang Y, Chen Y, Zhang J, et al. Cuproptosis: A novel therapeutic target for overcoming cancer drug resistance. Drug Resist Updat 2024;72:101018. [Crossref] [PubMed]
- Tang D, Kroemer G, Kang R. Targeting cuproplasia and cuproptosis in cancer. Nat Rev Clin Oncol 2024;21:370-88. [Crossref] [PubMed]
- Tong X, Tang R, Xiao M, et al. Targeting cell death pathways for cancer therapy: recent developments in necroptosis, pyroptosis, ferroptosis, and cuproptosis research. J Hematol Oncol 2022;15:174. [Crossref] [PubMed]
- Chen Y, Li X, Sun R, et al. A broad cuproptosis landscape in inflammatory bowel disease. Front Immunol 2022;13:1031539. [PubMed]
- Cai Y, He Q, Liu W, et al. Comprehensive analysis of the potential cuproptosis-related biomarker LIAS that regulates prognosis and immunotherapy of pan-cancers. Front Oncol 2022;12:952129. [PubMed]
- Yi X, Kim K, Yuan W, et al. Mice with heterozygous deficiency of lipoic acid synthase have an increased sensitivity to lipopolysaccharide-induced tissue injury. J Leukoc Biol 2009;85:146-53. [PubMed]
- Landgraf BJ, McCarthy EL, Booker SJ. Radical S-Adenosylmethionine Enzymes in Human Health and Disease. Annu Rev Biochem 2016;85:485-514. [Crossref] [PubMed]
- Burr SP, Costa AS, Grice GL, et al. Mitochondrial Protein Lipoylation and the 2-Oxoglutarate Dehydrogenase Complex Controls HIF1α Stability in Aerobic Conditions. Cell Metab 2016;24:740-52. [PubMed]
- Fan J, Bellon M, Ju M, et al. Clinical significance of FBXW7 loss of function in human cancers. Mol Cancer 2022;21:87. [PubMed]


