
circFADS2 inhibits ferroptosis associated with IGF2BP2-dependent SLC7A11 m6A modification in colorectal cancer cells
Highlight box
Key findings
• Our study demonstrates the molecular mechanism by which circFADS2 inhibits ferroptosis in colorectal cancer (CRC) cells by regulating IGF2BP2-mediated SLC7A11 m6A modification.
What is known and what is new?
• Studies have shown that inducing ferroptosis can have a good synergistic effect in tumor treatment.
• We explored the mechanism of action by knocking down circFADS2 and overexpressing SLC7A11 in CRC cell lines HT29 and HCT116, suggesting a possible therapeutic method for the clinical treatment of CRC.
What is the implication, and what should change now?
• Our findings offer fresh perspectives on the molecular process underlying CRC ferroptosis and lay a scientific foundation for developing new treatment strategies.
Introduction
Colorectal cancer (CRC) is a common cancer, the its incidence rate and mortality of which among young people are rising (1). Although the therapeutic approaches for CRC have been improved, symptoms of CRC only appear in late stages, and many patients are diagnosed with late-stage CRC at their first visit, resulting in relatively low cure rates and 5-year survival rates for patients (2). Therefore, understanding the potential molecular mechanisms underlying CRC is of enormous importance for developing novel treatment methods.
Iron-induced cell death (ferroptosis) is a cell death pattern distinct from apoptosis, which occurs due to the accumulation of iron or the consumption of antioxidant glutathione (GSH), leading to an increase in lipid peroxidation during steady-state turnover (3). Ferroptosis can inhibit tumor progression under certain circumstances (4); N6 methyladenosine modification enhances the anti ferroptotic ability of hepatoblastoma by inhibiting the deacetylation of SLC7A11 messenger RNA (mRNA), a member of the solute carrier family 7 (5). SLC7A11 is crucial for the uptake of cysteine, synthesis of GSH, and inhibition of ferroptosis (6). SLC7A11 is considered to play an important role in the ferroptosis process of tumor cells (7-9). Downregulation of SLC7A11 can promote ferroptosis in tumor cells, thereby exerting tumor suppressive functions (10). Existing studies have suggested that inducing ferroptosis may have a synergistic effect in tumor treatment (11,12). Therefore, exploring the mechanism of SLC7A11-induced ferroptosis may provide new insights for the development of CRC treatment strategies.
Circular RNA (circRNA) is mainly produced through a special alternative splicing method and is a non-coding RNA molecule (13). Currently, circRNA is known to play a role in various physiological processes (14), and also regulates gene expression by binding to specific mRNA (15). CircMAP2K4 upregulates YTHDF1 through m6A RNA methylation, thereby promoting cancer progression (16). According to a previous study, increased expression of circFADS2 is associated with lower overall survival rates in cancer patients (17). In addition, circFADS2 is the most significantly upregulated circRNA in CRC tissue, and its upregulation is closely related to the size, depth of invasion, metastasis, and poor prognosis of CRC (18). Therefore, we speculate that circFADS2 may play a role in CRC by regulating mRNA methylation modification.
M6A is considered one of the most abundant base modifications in eukaryotic mRNA, involved in biological processes such as mRNA modification and stability (19). According to reports, IGF2BP2 is an m6A reader that enhances mRNA stability by recognizing m6A modification sites (20). IGF2BP2 has been shown to promote cancer progression by enhancing the stability of SLC7A11 mRNA (21). Promoting SLC7A11 expression can enhance GPX4 activity to inhibit ferroptosis in CRC cells, thereby promoting CRC tumorigenesis (11). This suggests that regulating IGF2BP2-mediated SLC7A11 m6A modification may become an effective pathway for CRC treatment.
We speculated that circFADS2 may inhibit ferroptosis in CRC cells by regulating IGF2BP2-mediated SLC7A11 m6A modification. At present, there is no research to support the regulatory effect of circFADS2 on SLC7A11. We explored the mechanism of action by knocking down circFADS2 and overexpressing SLC7A11 in CRC cell lines HT29 and HCT116, which may provide support for the treatment of CRC. We present this article in accordance with the ARRIVE and MDAR reporting checklists (available at https://jgo.amegroups.com/article/view/10.21037/jgo-2024-1014/rc).
Methods
Cell culture and processing
We prepared a protocol before the study without registration. CRC cell lines HCT116 and HT29 were purchased from Cytion (Eppelheim, Germany). Cells were cultivated in Dulbecco’s modified Eagle medium (DMEM, 11965092, Gibco, Waltham, MA, USA) containing 10% fetal bovine serum (FBS, 26140079, Gibco) and 1% penicillin streptomycin (15140122, Gibco) at 37 ℃ and 5% CO2. When the cells were cultured to a confluence of 60–80%, lentiviral particles (purchased from VectorBuilder, Chicago, IL, USA) containing knockdown gene expression vectors (sh-circFADS2 1 #, sh-circFADS2 2 #, and sh-circFADS2 3 #) and overexpression gene expression vectors (oe-NC, oe-SLC7A11) were added for 48 hours, and antibiotics were added for cell screening.
Reverse transcription quantitative PCR
Total RNA was extracted using the TRIzol reagent (15596026, Invitrogen™, Waltham, MA, USA). Next, the extracted RNA was then quantified using a HD-UV90 spectrophotometer (Shandong Hold Electronic Technology Co., Weifang, China) as directed in the manual. After that, 2 µg of RNA was subjected to reverse transcription with the Vazyme DLR102 SynScript® III One-Step RT Kit (DLR102, Vazyme Biotech Co., Ltd., Nanjing, China) and microRNA (miRNA) First Strand cDNA Synthesis Kit (B532451, Sangon Biotech Co., Ltd., Shanghai, China) to create complementary DNA (cDNA). Later, the quantitative polymerase chain reaction (qPCR) analysis was performed in compliance with the instructions of the TB Green® Premix Ex Taq™ II (RR820A, Takara Holdings Inc., Kyoto, Japan). Subsequently, using GAPDH as an internal reference, the relative expression level (22) was calculated using the 2-ΔΔCt method. The primer sequences are shown in Table 1.
Table 1
| Gene | Primer sequences (5'-3') |
|---|---|
| circFADS2 | Forward: TTTGTGTGTGCGTGTTGTTGG |
| Reverse: ATGGAGGAGGCTTGGACATC | |
| SLC7A11 | Forward: TCCTGCTTTGGCTCCATGAACG |
| Reverse: AGAGGAGTGTGCTTGCGGACAT | |
| U6 | Forward: CTCGCTTCGGCAGCACAT |
| Reverse: TTTGCGTGTCATCCTTGCG | |
| GAPDH | Forward: GTCTCCTCTGACTTCAACAGCG |
| Reverse: ACCACCCTGTTGCTGTAGCCAA |
RT-qPCR, reverse transcription quantitative polymerase chain reaction.
Cell Counting Kit-8
A Cell Counting Kit-8 detection kit (C0037, Beyotime, Shanghai, China) was used for testing. Cells were inoculated (3×103/well) onto a 96 well plate and incubated. After 0, 12, 24, and 48 hours, cell samples were cultured at 37 ℃ for 2 hours with 10 µL CCK-8 and 90 µL serum-free medium, containing 95% air and 5% carbon dioxide. The optical density (OD) value was measured at a wavelength of 450 nm using a microplate reader (BioTek Instruments Inc, Winooski, VT, USA) to evaluate cell proliferation.
Colony formation assay
The cell suspension was diluted and each group of cells was inoculated into a cell culture dish at a density of 1,000 cells per dish. The cells were cultivated in a 37 ℃ 5% CO2 and saturated humidity incubator for 2 weeks. The supernatant was discarded and the cells were fixed with 4% paraformaldehyde (158127, Sigma Aldrich, St. Louis, MO, USA) for 15 minutes. Then, the fixative was removed, 10% diluted Giemsa staining solution (48900, Sigma Aldrich) was added for 10 minutes, the staining solution was washed away, and the cells were air dried. Clones with more than 10 cells were counted under a microscope to evaluate cell proliferation.
Transwell
The upper chamber of the Transwell was seeded with 200 μL of serum-free DMEM containing 1×105 cells (A1896702, Thermo Fisher Scientific, Waltham, MA, USA), while the lower chamber contained 500 μL of DMEM medium supplemented with 10% FBS. After co-culturing for 48 hours, the upper chamber cells were removed, the lower chamber cells were fixed with 4% paraformaldehyde, stained with crystal violet solution (198099, Merck, Darmstadt, Germany), and finally observed and photographed under a Nikon Eclipse E200 microscope (Nikon Corp., Tokyo, Japan). Quantitative analysis of migrating cells was performed using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Reactive oxygen species
Cells were digested with trypsin, the cell pellet was resuspended in 1 mL of pre-diluted 2',7'-dichlorodihydrofluorescein diacetate (DCFH-DA) probe (C400, Thermo Fisher Scientific) solution (working concentration of 10 µM), and left to sit at 37 ℃ in the dark for 30 minutes, with vortexing performed every 5 minutes, followed by centrifugation at 1,000 g and 25 ℃ for 5 minutes. Fluorescence images were obtained using a fluorescence microscope (Olympus, Tokyo, Japan) and analyzed using ImageJ software.
Terminal deoxynucleotidyl transferase dUTP nick end labeling
Trypsin was used to digest cells. The cells were fixed in a 4% formaldehyde solution for 20 minutes, then 0.1% Triton X-100 was added and incubated at room temperature for 5 minutes to allow permeabilization. The reaction solution was prepared by combining terminal deoxynucleotidyl transferase (TdT) with Alexa Fluor™ 488-dUTP: prepare the reaction solution according to the instructions of the terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) kit (C10617, Thermo Fisher Scientific). The reaction solution was added dropwise onto the sample and incubated at 37 ℃ for 1 hour. 4’,6-diamidino-2-phenylindole (DAPI) staining solution (62248, Thermo Fisher Scientific) was then added dropwise onto the sample and incubated at room temperature in the dark for 20 minutes. Apoptotic cells were observed under a fluorescence microscope, and the apoptosis rate was determined by counting the total number of cells and the number of apoptotic cells (apoptosis rate = TUNEL positive cells/total cells * 100%).
Western blot (WB)
Radioimmunoprecipitation assay (RIPA) solution (89900, Thermo Fisher Scientific) was added to the cells which were then lysed on ice for 30 minutes and shaken every 5 minutes during this period. They were then centrifuged at 12,000 rpm for 10 minutes at 4 ℃ and the supernatant was collected. The protein concentration of each sample was measured using a bicinchoninic acid (BCA) protein detection kit (23227, Thermo Fisher Scientific). After separation with sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), the protein was transferred to a polyvinylidene fluoride (PVDF) membrane (88518, Thermo Fisher Scientific). After electroporation, the membrane was sealed with 5% skimmed milk powder for 1 hour. Following blocking, SLC7A11 (1:1,000, PA1-16893, Thermo Fisher Scientific) and GAPDH (1:5,000, MA5-35235, Invitrogen) antibodies were added to treat the membrane, and incubated overnight at 4 ℃. Then, goat anti rabbit IgG HRP antibody diluted in 5% skim milk (1:10,000, 31460, Thermo Fisher Scientific) was added, followed by incubation at room temperature for 1 hour. Finally, the protein bands were developed using enhanced chemiluminescence (ECL) reagent (32106, Thermo Fisher Scientific) and analyzed for OD using ImageJ image analysis software with GAPDH as the internal reference.
Biochemical analysis
A reduced GSH Colorimetric Assay Kit (EIAGSHC, Thermo Fisher Scientific), MDA Kit (HY-K0319, MedChemExpress, Monmouth Junction, NJ, USA) and cell total iron colorimetric assay kit (EEA009, Thermo Fisher Scientific) was used to detect GSH, MDA, ROS, and Fe2+ in cells.
Animal modeling and processing
This study was approved by the Animal Committee of Affiliated Nanhua Hospital, University of South China (No. 20240302) and conducted in compliance with the national guidelines for the care and use of animals. The supplier of the 6-week-old male BALB/c nude mice (16–18 g) was Fujian Anbuli Biotechnology Co., Ltd. (Fujian, China). All animals were kept in a pathogen-free environment and were free to eat and drink in a 12-hour light/dark cycle, with room temperature of 21±2 ℃ and humidity ranging from 45% to 65%. Only healthy, 6-week-old male BALB/c nude mice without prior medical history were selected for the experiment. Twenty-four BALB/c nude mice were randomly divided into 4 groups (n=6): sh-NC, sh-circFADS2, sh-circFADS2 + oe-NC, and sh-circFADS2 + oe-SLC7A11. A subcutaneous injection of 1×106 stably transfected HT29 cells [suspended in 0.2 mL phosphate-buffered saline (PBS)] was administered into the right abdomen of BALB/c nude mice. After 7 days of vaccination, the long and short axes of the tumor were measured every 4 days, and the tumor volume (V) was calculated using the formula (V=1/2 × long axis × short axis2). After the experiment was completed (day 28), the tumor volume was measured and the mice were euthanized via intraperitoneal injection of an excess (200 mg/kg) of pentobarbital sodium (57-33-0, Merck, Darmstadt, Germany). The tumor tissue was removed and weighed.
RNA immunoprecipitation
After the cells had been cultivated to a confluence degree of 70–80%, the culture medium was removed and RIPA buffer containing protease and RNAse inhibitors was added for lysis for 30 minutes. Following centrifugation at 12,000 rpm for 15 minutes, the supernatant was collected. Incubation was performed with protein A/G magnetic beads with lysis buffer for 30 minutes, followed by washing with PBS. Then, IGF2BP2 antibody (712137, Thermo Fisher Scientific) and IgG antibody (SAB5600195, Sigma Aldrich) were added and incubated overnight at 4 ℃, after which protein A/G beads (88802, Thermo Fisher Scientific) were added. The mixture was rotated and incubated for 2 hours to allow the antibody antigen complex to bind to the beads. The beads were then washed 3 times and centrifuged each time to remove the supernatant. Subsequently, proteinase K (20 mg/mL) was added for cross-linking. The purified RNA was extracted and analyzed by RT-qPCR.
Nuclear cytoplasmic separation experiment
SurePrep™ Nuclear or Cytolytic RNA Purification Kit (BP2805-50, Thermo Fisher Scientific) was used to treat cells with cell lysis buffer according to the instructions in the kit. The cells were then incubated on ice for 5–10 minutes to lyse the cell membrane and release cytoplasmic contents. According to the instructions of the reagent kit, nuclear extraction buffer was used to treat nuclear pellet. Following centrifugation at 10,000 ×g at 4 ℃ for 20 minutes, the supernatant was collected and precipitated for RT-qPCR analysis.
mRNA stability testing
Cells that had undergone knockdown of circFADS2 and IGF2BP2, as well as negative control cells, were treated with 1 µg/mL transcription inhibitor Actinomycin D (J60148.LB0, Thermo Fisher Scientific) to block the synthesis of new mRNA. In order to assess the stability of the expression of SLC7A11 mRNA, cells were collected at various time points (0, 1, 2, and 4 h) and tested by RT-qPCR.
Fluorescence in situ hybridization (FISH) and immunofluorescence
A FISH kit purchased from Sigma Aldrich was used to measure the fluorescence signal of the probe, and fluorescence image acquisition was completed using a Zeiss LSM 980 microscope (ZEISS, Oberkochen, Germany). Cells were fixed with 4% formaldehyde and incubated at room temperature for 30 minutes. Cell permeabilization was performed with 0.5% Triton X-100 treatment. The cell samples were mixed with 50 nM fluorescein isothiocyanate (FITC)-labeled circFADS2 and SLC7A11 mRNA probes purchased from Sigma Aldrich, and incubated at 37 ℃ for 4 hours in a wet box.
Immunofluorescence detection: cells were fixed with 4% formaldehyde and incubated at room temperature for 30 minutes. Cell permeabilization was performed using 0.5% Triton X-100 treatment. The sample was washed with pre-cooled saline-sodium citrate (SSC) buffer and IGF2BP2 antibody was added (1:100, PA5-115396, Thermo Fisher Scientific) for overnight incubation at 4 ℃. Goat anti rabbit IgG H&L (Alexa Fluor® 647) (1 µg/mL, ab150079, Abcam, Cambridge, MA, USA) was added and incubated at room temperature in the dark for 2 hours. Then, 1 µg/mL DAPI (62248, Thermo Fisher Scientific) and incubate for 10 minutes. After sealing with DAPI Aqueous, Fluoroshield (ab104139, Abcam), the cells were viewed under a fluorescent microscope. ImageJ image analysis software was used to analyze the fluorescence images.
Statistical analysis
GraphPad Prism 9 (GraphPad Software, San Diego, CA, USA) was used for statistical analysis. Comparisons between 2 groups were performed using the t-test. For data involving 3 or more groups, 1- or 2-way analysis of variance (ANOVA) was used, followed by Tukey’s post hoc test. A P value <0.05 indicated a statistical difference, and at least 3 replications of each experiment were conducted.
Results
Knocking down circFADS2 inhibits malignant behavior of CRC cells
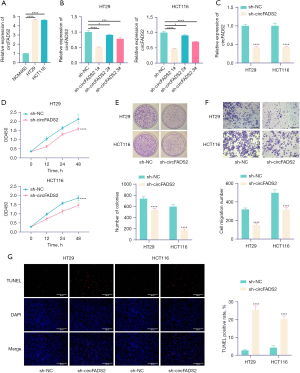
RT-qPCR analysis revealed that circFADS2 was upregulated in CRC cells (HT29 and HCT116) (Figure 1A). Knockdown of circFADS2 was performed on CRC cells, and circFADS2 expression was detected by RT-qPCR. The outcomes demonstrated that the downregulation of circFADS2 was most significant in sh-circFADS2 1 # (Figure 1B). Based on the experimental results, we selected the infection of sh-circFADS2 1 # for subsequent experiments. Knocking down circFADS2 obviously led to downregulation of circFADS2 expression (Figure 1C). CCK-8 detection showed that knocking down circFADS2 significantly reduced CRC cell proliferation (Figure 1D). The colony formation assay showed that downregulating the expression of circFADS2 inhibited the ability of CRC cells to form clones (Figure 1E). The Transwell findings demonstrated that the migration ability of CRC cells decreased after the expression of circFADS2 was reduced (Figure 1F). TUNEL detection showed that knockdown of circFADS2 promoted apoptosis of CRC cells (Figure 1G). The outcomes demonstrated that knocking down circFADS2 can inhibit the malignant behavior of CRC cells.
Knocking down circFADS2 promotes ferroptosis in CRC cells
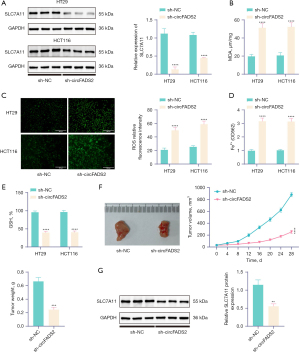
WB detection showed that knocking down circFADS2 significantly reduced the expression of ferroptosis-related protein SLC7A11 in CRC cells (Figure 2A). In addition, after circFADS2 knockdown, the GSH level in CRC cells significantly decreased, while the concentrations of MDA, ROS, and Fe2+ increased significantly (Figure 2B-2E). In BALB/c nude mice, CRC cells knocking down circFADS2 were injected subcutaneously into the lower abdomen of the mice. In addition, knocking out circFADS2 significantly reduced tumor growth and tumor weight in BALB/c mice (Figure 2F). The WB detection results showed that compared with the sh-NC group, the expression of ferroptosis related protein SLC7A11 was reduced in the tumor tissues of BALB/c nude mice in the sh-circFADS2 group (Figure 2G). The results indicate that knocking down circFADS2 promotes ferroptosis in CRC cells.
The formation of stable complex circFADS2/IGF2BP2/SLC7A11 promotes the stability of SLC7A11 mRNA
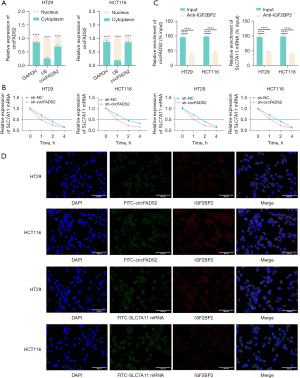
The findings from nuclear cytoplasmic separation experiments indicated that circFADS2 is mainly expressed in the cytoplasm (Figure 3A). RT-q-PCR detection showed that knocking down circFADS2 significantly reduced the stability of SLC7A11 mRNA. In addition, inhibiting IGF2BP2 was also shown to reduce the stability of SLC7A11 mRNA and decrease SLC7A11 expression (Figure 3B). IGF2BP2 is an m6A reader that enhances mRNA stability by recognizing m6A modification sites (20). The RNA immunoprecipitation (RIP) results indicated that there are interactions between circFADS2 and IGF2BP2, as well as between IGF2BP2 and SLC7A11 mRNA (Figure 3C). The FISH and IF analysis results showed that circFADS2 and IGF2BP2 were co-localized in the cytoplasm of CRC cells, and SLC7A11 mRNA was co-localized with IGF2BP2 in the cytoplasm (Figure 3D). The results indicated that circFADS2 promotes m6A methylation modification of SLC7A11 mRNA by IGF2BP2 through binding, thereby promoting SLC7A11 expression.
circFADS2 promotes malignant behavior of CRC cells by regulating SLC7A11
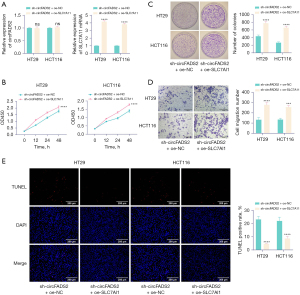
Overexpression of SLC7A11 was performed in circFADS2 knockdown CRC cells (HT29 and HCT116). The results of RT-qPCR showed that compared with the sh-circFADS2 + oe-NC group, SLC7A11 was upregulated in the sh-circFADS2 + oe-SLC7A11 group (Figure 4A). CCK-8 detection showed that after upregulation of SLC7A11, CRC cell proliferation significantly increased (Figure 4B). The colony formation assay showed that further expression of SLC7A11 increased CRC cell proliferation (Figure 4C). The Transwell results indicated that overexpression of SLC7A11 promoted CRC cell migration (Figure 4D). TUNEL detection showed that overexpression of SLC7A11 inhibited apoptosis in CRC cells (Figure 4E). The results showed that overexpression of SLC7A11 reversed the inhibitory effect of knocking down circFADS2 on the malignant behavior of CRC cells. This indicates that circFADS2 promotes malignant behavior of CRC cells by regulating SLC7A11.
circFADS2 inhibits ferroptosis in CRC cells by regulating SLC7A11 expression
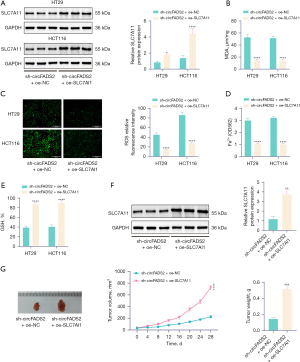
In the in vitro experiments, WB detection showed that overexpression of SLC7A11 significantly increased SLC7A11 expression in CRC cells (Figure 5A). In addition, overexpression of SLC7A11 significantly increased GSH levels in CRC cells, whereas MDA, ROS levels, and Fe2+ concentrations were significantly reduced (Figure 5B-5E). CRC cells overexpressing SLC7A11 were injected subcutaneously into the lower abdomen of BALB/c nude mice. WB detection showed that compared with the sh-circFADS2 + oe-NC group, the sh-circFADS2 + oe-SLC7A11 group showed an increase in SLC7A11 expression in BALB/c nude mice (Figure 5F). After promoting SLC7A11 expression, tumor growth and tumor weight significantly increased in BALB/c nude mice (Figure 5G). The results showed that overexpression of SLC7A11 reversed the effect of knocking down circFADS2 on promoting ferroptosis in CRC cells, indicating that circFADS2 inhibits ferroptosis in CRC cells by regulating SLC7A11 expression.
Discussion
CRC is the second deadliest cancer worldwide (23). Although improvements have been made in CRC treatment, the treatment and prognosis are still not ideal (24). Therefore, understanding the molecular mechanisms underlying the onset of CRC is crucial for improving the poor prognosis of CRC patients.
CircRNAs are involved in many different physiological processes, and many circRNAs are differentially expressed in CRC and matched normal tissues (25). circFADS2 is the most significantly upregulated circRNA in CRC tissue (18). Therefore, we validated its role in CRC by knocking down circFADS2. We have demonstrated in our research that circFADS2 plays an important role in CRC cells. After knocking down circFADS2, the cell proliferation and migration ability of CRC cells significantly decreased, whereas apoptosis significantly increased. This suggests that circFADS2 may serve as a new breakthrough point for the treatment of CRC.
During ferroptosis, ROS production increases, mitochondrial volume decreases, and membrane density increases (26). It is often associated with lipid peroxidation caused by increased GSH consumption or lipid peroxidation, as well as SLC7A11 deficiency (27). Ferroptosis has been shown to inhibit tumors (28). Our research results indicate that downregulating circFADS2 significantly reduces the expression of SLC7A11 in CRC cells. At the same time, the ROS and MDA levels in the cells were significantly reduced, the Fe2+ concentration was decreased, and the GSH level was increased. It was made clear that knocking down circFADS2 promotes ferroptosis in CRC cells.
As a key regulatory protein of ferroptosis, SLC7A11 has been reported to promote CRC progression (7). In addition, a study has shown that m6A methylation of SLC7A11 mRNA plays a key role in the regulation of ferroptosis in CRC (29). IGF2BP2, as an m6A reader, can enhance mRNA stability by recognizing m6A modification sites (20). In oral squamous cell carcinoma, METTL3 enhances the stability of SLC7A11 mRNA through m6A-mediated binding of IGF2BP2 (21). To verify the mechanism by which circFADS2 regulates SLC7A11, we demonstrated the interaction between IGF2BP2 and circFADS2, as well as between IGF2BP2 and SLC7A11 mRNA, through RIP. It was demonstrated through nuclear cytoplasmic separation that circFADS2 is mainly expressed in the cytoplasm. In addition, through stability testing of SLC7A11 mRNA, we found that overexpression of circFADS2 or increase of IGF2BP2 can improve the stability of SLC7A11 mRNA. We infer that circFADS2 may regulate the stability of SLC7A11 mRNA through IGF2BP2.
Afterwards, we demonstrated the co localization of circFADS2, IGF2BP2, and SLC7A11 mRNA in the cytoplasm through IF-FISH analysis. Here, we provide an effective mechanism by which circFADS2 enhances the stability of SLC7A11 mRNA by promoting the formation of the circFADS2/IGF2BP2/SLC7A11 mRNA protein complex. Although a previous study has shown that circFADS2 is the most significantly upregulated circular RNA in CRC tissues (18), no research has investigated the role and regulatory mechanism of circFADS2 in CRC ferroptosis. Furthermore, we acknowledge that although we have demonstrated in vitro and in vivo that circFADS2 inhibits ferroptosis in CRC cells by regulating IGF2BP2-mediated SLC7A11 m6A modification, in vitro CRC cell lines and in vivo mouse models may not fully replicate all pathological features and immune responses of human CRC. This may affect the translatability of research results.
Our study demonstrates the molecular mechanism by which circFADS2 inhibits ferroptosis in CRC cells by regulating IGF2BP2-mediated SLC7A11 m6A modification. This offers fresh perspectives on the molecular process underlying CRC ferroptosis and lays a scientific foundation for developing new treatment strategies.
Conclusions
We have demonstrated that circFADS2 can promote the formation of the circFADS2/IGF2BP2/SLC7A11 mRNA protein complex, thereby promoting SLC7A11 m6A methylation. This clearly promotes ferroptosis in CRC cells. This offers a fresh line of inquiry for CRC therapy and theoretical support for improving its poor prognosis.
Acknowledgments
None.
Footnote
Reporting Checklist: The authors have completed the ARRIVE and MDAR reporting checklists. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-2024-1014/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-2024-1014/dss
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-2024-1014/prf
Funding: This study was supported by
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-2024-1014/coif). The authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bhandari A, Woodhouse M, Gupta S. Colorectal cancer is a leading cause of cancer incidence and mortality among adults younger than 50 years in the USA: a SEER-based analysis with comparison to other young-onset cancers. J Investig Med 2017;65:311-5. [Crossref] [PubMed]
- Liau XL, Salvamani S, Gunasekaran B, et al. CCAT 1- A Pivotal Oncogenic Long Non-Coding RNA in Colorectal Cancer. Br J Biomed Sci 2023;80:11103. [Crossref] [PubMed]
- Ye S, Xu M, Zhu T, et al. Cytoglobin promotes sensitivity to ferroptosis by regulating p53-YAP1 axis in colon cancer cells. J Cell Mol Med 2021;25:3300-11. [Crossref] [PubMed]
- Chen D, Chu B, Yang X, et al. iPLA2β-mediated lipid detoxification controls p53-driven ferroptosis independent of GPX4. Nat Commun 2021;12:3644. [Crossref] [PubMed]
- Liu L, He J, Sun G, et al. The N6-methyladenosine modification enhances ferroptosis resistance through inhibiting SLC7A11 mRNA deadenylation in hepatoblastoma. Clin Transl Med 2022;12:e778. [Crossref] [PubMed]
- Wang X, Chen Y, Wang X, et al. Stem Cell Factor SOX2 Confers Ferroptosis Resistance in Lung Cancer via Upregulation of SLC7A11. Cancer Res 2021;81:5217-29. [Crossref] [PubMed]
- Shen L, Zhang J, Zheng Z, et al. PHGDH Inhibits Ferroptosis and Promotes Malignant Progression by Upregulating SLC7A11 in Bladder Cancer. Int J Biol Sci 2022;18:5459-74. [Crossref] [PubMed]
- Yang J, Zhou Y, Xie S, et al. Metformin induces Ferroptosis by inhibiting UFMylation of SLC7A11 in breast cancer. J Exp Clin Cancer Res 2021;40:206. [Crossref] [PubMed]
- Chen Q, Zheng W, Guan J, et al. SOCS2-enhanced ubiquitination of SLC7A11 promotes ferroptosis and radiosensitization in hepatocellular carcinoma. Cell Death Differ 2023;30:137-51. [Crossref] [PubMed]
- Jiang L, Kon N, Li T, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature 2015;520:57-62. [Crossref] [PubMed]
- Qiao Y, Su M, Zhao H, et al. Targeting FTO induces colorectal cancer ferroptotic cell death by decreasing SLC7A11/GPX4 expression. J Exp Clin Cancer Res 2024;43:108. [Crossref] [PubMed]
- Wang H, Breadner DA, Deng K, et al. CircRHOT1 restricts gastric cancer cell ferroptosis by epigenetically regulating GPX4. J Gastrointest Oncol 2023;14:1715-25. [Crossref] [PubMed]
- Yao B, Zhang Q, Yang Z, et al. CircEZH2/miR-133b/IGF2BP2 aggravates colorectal cancer progression via enhancing the stability of m(6)A-modified CREB1 mRNA. Mol Cancer 2022;21:140. [Crossref] [PubMed]
- Wang F, Nazarali AJ, Ji S. Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am J Cancer Res 2016;6:1167-76. [PubMed]
- Xia H, Wu Y, Zhao J, et al. N6-Methyladenosine-modified circSAV1 triggers ferroptosis in COPD through recruiting YTHDF1 to facilitate the translation of IREB2. Cell Death Differ 2023;30:1293-304. [Crossref] [PubMed]
- Chi F, Cao Y, Chen Y. Analysis and Validation of circRNA-miRNA Network in Regulating m(6)A RNA Methylation Modulators Reveals CircMAP2K4/miR-139-5p/YTHDF1 Axis Involving the Proliferation of Hepatocellular Carcinoma. Front Oncol 2021;11:560506. [Crossref] [PubMed]
- Zhao F, Han Y, Liu Z, et al. circFADS2 regulates lung cancer cells proliferation and invasion via acting as a sponge of miR-498. Biosci Rep 2018;38:BSR20180570. [Crossref] [PubMed]
- Xiao YS, Tong HZ, Yuan XH, et al. CircFADS2: A potential prognostic biomarker of colorectal cancer. Exp Biol Med (Maywood) 2020;245:1233-41. [Crossref] [PubMed]
- Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol 2017;18:31-42. [Crossref] [PubMed]
- Hou P, Meng S, Li M, et al. LINC00460/DHX9/IGF2BP2 complex promotes colorectal cancer proliferation and metastasis by mediating HMGA1 mRNA stability depending on m6A modification. J Exp Clin Cancer Res 2021;40:52. [Crossref] [PubMed]
- Xu L, Li Q, Wang Y, et al. m(6)A methyltransferase METTL3 promotes oral squamous cell carcinoma progression through enhancement of IGF2BP2-mediated SLC7A11 mRNA stability. Am J Cancer Res 2021;11:5282-98. [PubMed]
- Shen M, Cao S, Long X, et al. DNAJC12 causes breast cancer chemotherapy resistance by repressing doxorubicin-induced ferroptosis and apoptosis via activation of AKT. Redox Biol 2024;70:103035. [Crossref] [PubMed]
- Long F, Li L, Xie C, et al. Intergenic CircRNA Circ_0007379 Inhibits Colorectal Cancer Progression by Modulating miR-320a Biogenesis in a KSRP-Dependent Manner. Int J Biol Sci 2023;19:3781-803. [Crossref] [PubMed]
- Mi X, Yao H, Lu Y, et al. Leptin increases chemosensitivity by inhibiting CPT1B in colorectal cancer cells. J Gastrointest Oncol 2024;15:2507-20. [Crossref] [PubMed]
- Zhou J, Wang L, Sun Q, et al. Hsa_circ_0001666 suppresses the progression of colorectal cancer through the miR-576-5p/PCDH10 axis. Clin Transl Med 2021;11:e565. [Crossref] [PubMed]
- Jiang X, Stockwell BR, Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol 2021;22:266-82. [Crossref] [PubMed]
- He F, Zhang P, Liu J, et al. ATF4 suppresses hepatocarcinogenesis by inducing SLC7A11 (xCT) to block stress-related ferroptosis. J Hepatol 2023;79:362-77. [Crossref] [PubMed]
- Li D, Wang Y, Dong C, et al. CST1 inhibits ferroptosis and promotes gastric cancer metastasis by regulating GPX4 protein stability via OTUB1. Oncogene 2023;42:83-98. [Crossref] [PubMed]
- Qiao Y, Su M, Zhao H, et al. Correction: Targeting FTO induces colorectal cancer ferroptotic cell death by decreasing SLC7A11/ GPX4 expression. J Exp Clin Cancer Res 2024;43:131. [Crossref] [PubMed]


