TNFRSF10C copy number variation is associated with metastatic colorectal cancer
Introduction
Colorectal cancer (CRC) is the third leading cause of cancer-related death in the United States in both men and women, and approximately 20% of all current patients with CRC have distant metastatic disease (1) Despite advances in screening and treatment, the 5-year overall survival for patients with CRC is still dismal (1). Genes associated with the presence and development of CRC and markers for drug response have been characterized (2-4), but genetic markers associated with metastatic disease in CRC are not well defined. Treatment strategies for CRC are dependent upon stage and location of the primary tumor, and may use a combination of surgical resection, chemotherapy, and radiation therapy. In particular, radiation therapy improves local control and overall survival in patients with rectal adenocarcinoma (5). Copy number variations (CNVs) have been used to determine prognosis and subsequent treatment profile for a number of cancers, e.g., MYCN amplification for neuroblastoma (6). Comparisons between CNV of genes in primary CRC tumors and their matched metastatic disease sites are well documented, but less is known about how CNV of genes in CRC primary tumors differs between those associated with localized disease compared with those associated with distant metastatic disease (7-12).
Prior studies have shown a role for TNF-related apoptosis inducing ligand (TRAIL) dysregulation at multiple cancer sites, and a smaller study showed evidence of down regulation of tumor necrosis factor receptor superfamily member 10C (TNFRSF10C) at the sites of metastatic disease in CRC (13-16). TNFRSF10C, also known as decoy receptor-1 (DcR1) and TRAIL-R3, is a decoy TRAIL receptor, which functions as an antagonistic receptor that protects cells from TRAIL-induced apoptosis (17). TNFRSF10C expression is often down regulated in cancer (13-15), and loss of TNFRSF10C sensitizes cells to TRAIL-induced apoptosis (18). Indeed, TRAIL genes are prognostic for response to chemotherapy and overall survival in patients with CRC, and moreover are prognostic for outcome in patients with other cancers, including glioblastoma multiforme and breast cancer (19-24). Differences in TRAIL expression in cancers compared with normal cells have led to clinical trials targeting TRAIL-induced apoptosis in CRC, but the genetic profiles of patients were not used as part of the selection criteria (25-29).
Tri-modality treatment with surgery, chemotherapy, and radiation therapy represents the standard of care in the majority of patients with stage II-III rectal adenocarcinoma. The current PROSPECT clinical trial is examining the possible omission of radiation therapy in select patients with rectal adenocarcinoma and instead using more aggressive chemotherapy strategies (30). Identifying genetic markers that can discriminate whether patients have a propensity for localized vs. metastatic disease could lead to the individualization of treatment for patients with rectal adenocarcinoma and help to identify those patients who may be more likely to benefit from more aggressive chemotherapeutic strategies.
In this study, we assessed the association of TNFRSF10C CNV with distant metastatic disease in CRC using a cohort of 515 patients from The Cancer Genome Atlas (TCGA).
Methods
Patient data
A cohort of 515 samples taken from primary tumor specimen was selected from the TCGA database in April 2014 based on availability of both metastatic staging and copy number data. A subset of 144 samples with rectal primary and CNV data was separately analyzed. The results here are in part based upon data generated by the TCGA Research Network (http://cancergenome.nih.gov/) established by the NCI and NHGRI.
Clinicopathological data
Data on age, sex, staging, and race/ethnicity was collected from clinical information on the TCGA data portal. Pathologic findings of tumor size, tumor location, resection margins, lymph node status and metastatic status at diagnosis were also available through the TCGA data portal.
CNV analysis
The TCGA level 3 CNV data was extracted for colon and rectal adenocarcinoma from TCGA data portal. TCGA level 3 CNV (Affymetrix Genome-Wide SNP Array 6.0) was processed and normalized per sample. The mean copy number estimates of segments overlapping the whole genome were obtained and used for the analysis. Genomic identification of significant targets in cancer (GISTIC) algorithm mean cut-offs were used to categorize the gene. Copy numbers ≥1 or ≤−1 were defined as presence of CNV.
Statistical analysis
In the CRC set, univariate association of TNFRSF10C CNV with covariates was examined with chi-square test or Fisher’s exact test, where appropriate. Univariate analysis of metastatic disease (M1 vs. M0) with predictors and covariates was carried out with a logistic regression model. Multivariable analysis of metastatic disease (M1 vs. M0) was conducted with a logistic regression model by entering all variables in the model and using a backward variable selection method with an alpha level of removal of 0.1. TNFRSF10C CNV, chemotherapy, N stage, and T stage were forced in the model.
For the rectal subset, univariate association of TNFRSF10C CNV with covariates was examined with chi-square test or Fisher’s exact test, where appropriate. Univariate analysis of metastatic disease (M1 vs. M0) with the predictor and covariates was carried out with a logistic regression model. Multivariable analysis of metastatic disease (M1 vs. M0) was conducted with a logistic regression model by entering all variables in the model and using a backward variable selection method with an alpha level of removal of 0.1. TNFRSF10C CNV, N stage, and T stage were forced in the model. All analyses were done using SAS 9.3 (SAS Institute, Inc., Cary, North Carolina, USA) with a significant level of 0.05.
Upon determining significance between TNFRSF10C CNV and distant metastatic disease in both the colorectal cohort and rectal subset, TNFRSF10C CNV data was further queried to determine if the CNV was a gain or loss, and overwhelmingly the CNV was a homozygous deletion indicating copy number loss.
Results
The CRC copy number analysis cohort (Table 1) consisted of 515 patients with the diagnosis of CRC (270 male and 245 female). The median age at diagnosis was 68 years (range, 31–90 years). The rectal subset for copy number analysis (Table 2) contained 144 patients (77 male and 67 female) with a median age at diagnosis of 65 years (range, 31–90 years).
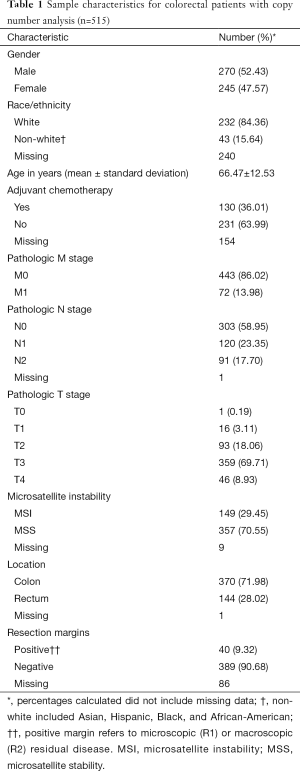
Full table
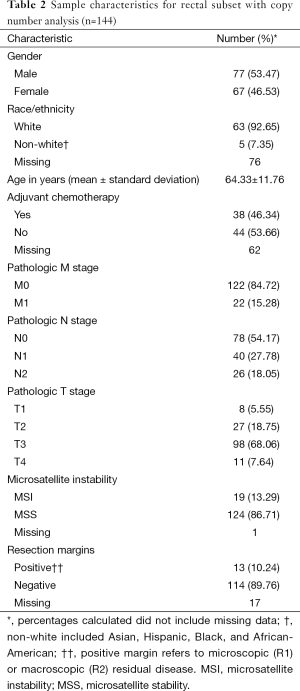
Full table
Pathological characteristics
In the CRC copy number analysis cohort, distant metastatic disease was diagnosed in 72 (14%) patients, and 211 (41%) patients had positive nodal disease. In the rectal subset, 22 (15%) patients had distant metastatic disease and 66 (46%) patients had positive nodal disease.
Genetic analysis for all patients
There were 27 samples that had CNV at TNFRSF10C, and the samples displayed decreased copy number through homozygous deletions. On univariate analysis, TNFRSF10 CNV demonstrated a statistically significant association with distant metastatic disease (P<0.001), positive nodal disease (P=0.005) and positive resection margins (P<0.001). There was no association found between TNFRSF10C CNV with microsatellite instability (MSI), location, or T stage (Table 3).
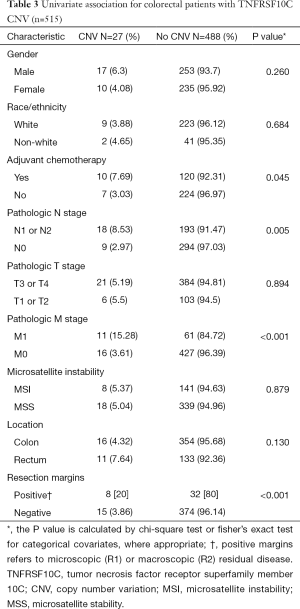
Full table
On univariate analysis for metastatic disease (Table 4), presence of distant metastatic disease was found to be associated with presence of TNFRSF10C CNV [odds ratio (OR) 4.81; 95% confidence interval (CI), 2.13–10.85; P<0.001], N1 or N2 nodal disease [OR 18.83 (95% CI, 8.42–42.09); P<0.001], T3 or T4 advanced local disease [OR 11.18 (95% CI, 2.70–46.36); P<0.001], positive resection margins [OR 90.14 (95% CI, 34.35–236.52); P<0.001], and use of adjuvant chemotherapy [OR 4.90 (95% CI, 2.54–9.44); P<0.001]. On multivariate analysis (Table 4), presence of TNFRSF10C CNV remained associated with presence of distant metastatic disease [OR 4.63 (95% CI, 1.31–16.43); P=0.018], in addition to N1 or N2 nodal disease [OR 14.45 (95% CI, 4.55–45.83); P<0.001]. Race/ethnicity, location, and resection margins were dropped from the multivariate model.
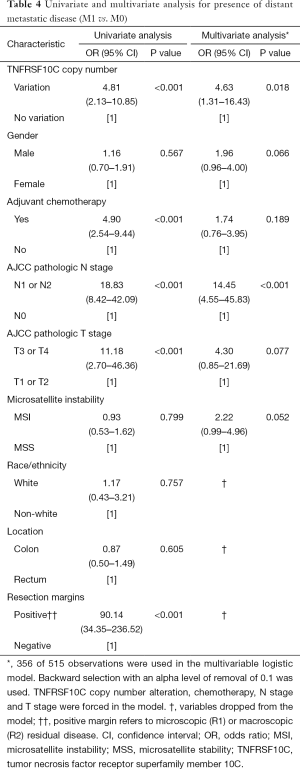
Full table
Subset analysis of rectal cancer patients
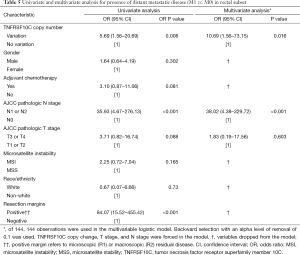
On univariate analysis for presence of metastatic disease in the rectal subset (Table 5), distant metastatic disease was significantly associated with TNFRSF10C CNV [OR 5.69 (95% CI, 1.56–20.69); P=0.008], N1 or N2 nodal disease [OR 35.93 (95% CI, 4.67–276.13); P<0.001] and positive resection margins [OR 84.07 (95% CI, 15.52–455.42); P<0.001]. Adjuvant chemotherapy, MSI and pathologic T stage were not associated with presence of distant metastatic disease. On multivariate analysis including TNFRSF10C CNV analysis, T stage, and N stage in the logistic regression model, presence of metastatic disease remained associated with TNFRSF10C alteration [OR 10.69 (95% CI, 1.56–73.15); P=0.016] and N1 or N2 nodal disease [OR 38.02 (95% CI, 4.38–329.72); P<0.001]. Adjuvant chemotherapy, gender, MSI, race/ethnicity, and resection margins were removed from the model.
Full table
Discussion
In this study, we found that TNFRSF10C CNV was independently associated with distant metastatic disease in CRC and the rectal subset from an analysis of data from 515 patients utilizing TCGA. We confirmed our findings on multivariate analysis, taking into account age, T stage, N stage, adjuvant chemotherapy, gender, MSI, location, and surgical margin status. Our findings provide evidence that TNFRSF10C CNV is significantly associated with distant metastatic disease in patients with all forms of CRC and in patients with rectal cancer. Additionally, these data suggest that CNV in TNFRSF10C may be a useful genetic marker to personalize treatment for patients with CRC, and potentially identify those patients who may benefit from more aggressive systemic treatment strategies.
A recent large-scale analysis of TCGA by Lee et al. did not find any evidence of a significant genetic association with distant metastatic disease when incorporating multiple genetic markers (mutations, CNV, gene expression, and methylation status) into the analysis (8). Our study differs in the finding of a CNV of a unique gene independently associated with distant metastatic disease utilizing a larger database with more samples having distant metastatic disease. In the Macartney-Coxson et al. study of 30 patients with CRC, low TNFRSF10C expression was associated with the development of extrahepatic metastases (13). Consistent with these findings, the instances of TNFRSF10C CNV in our cohort were overwhelmingly homozygous deletions, and therefore associated with downregulation of the gene. TNFRSF10C CNV is associated with more aggressive disease in CRC as demonstrated by its association with distant metastases and nodal disease in our cohort.
The current cohort contained many samples of both colectomy and metastatic disease thereby allowing correlative analysis with more statistical power. Distant metastatic disease is associated with nodal disease, and trends with advanced T stage. Another factor that also plays a role in the development of distant metastatic disease is the molecular pathway of tumorigenesis. Hereditary non-polyposis CRC is associated with left colon primary tumors with a lower propensity of forming metastases, as well as being associated with MSI (31) Several studies have indicated different molecular and clinical features amongst different locations in the colon (2,32,33), and for the aforementioned reasons we controlled for MSI when examining any association of TNFRSF10C CNV with distant metastatic disease. Although there was an expected trend between distant metastatic disease in the CRC cohort and microsatellite stability (MSS) (P=0.052) on multivariate analysis, TNFRSF10C CNV was not found to be associated with MSI/MSS on univariate analysis (P=0.879), and moreover, TNFRSF10C was independently associated with distant metastatic disease (P=0.018) on multivariate analysis. Taken together, these findings suggest that the relationship between TNFRSF10C CNV and distant metastatic disease is not dependent upon an association with the molecular differences between right-sided and left-sided primary tumors.
Variations in molecular expression and somatic changes have been reported between colon and rectal cancers (34,35). Due to these differences, subset analysis was performed in rectal adenocarcinoma samples. Rectal adenocarcinoma subset analysis confirmed that TNFRSF10C CNV was independently associated with distant metastases. Therefore, the reported changes between rectal and colon primaries do not seem to affect the association of TNFRSF10C CNV and distant metastases in rectal primary patients. Significantly, rectal and colon cancers have differences in treatment strategies. Radiation therapy is often used in rectal adenocarcinomas due to a higher incidence of local recurrence in rectal adenocarcinoma compared with CRC (36). Although local recurrence may be a main target in therapy for locally advanced tumors, there must also be consideration for the development of metastases as aggressive local treatment would be of less benefit. Multiple studies have shown that the number of lymph nodes sampled during surgery for CRC is correlated with prognosis in stage II CRC, thus implicating that further treatment may prevent a reservoir for the spread of cancer (37,38). With increasing evidence of spread of tumor in the early stages of CRC, the use of another indicator like TNFRSF10C CNV may be useful in guiding therapy toward a more aggressive regimen focused on preventing the spread of tumor. After surgery, rectal adenocarcinoma treatment will usually focus on a systemic therapy versus localized therapy, thus the use of a clinical marker associated with distant metastatic disease could help to influence treatment in rectal cancer to favor systemic therapy versus radiation therapy. The use of TNFRSF10C as a molecular marker to influence therapy will depend upon future studies having a longitudinal design thereby allowing more association with development of advanced disease with distant metastases.
We acknowledge that this study does have limitations in addition to the retrospective design. The data was obtained from patients who had resection of primary tumor, thereby not involving advanced, unresectable disease in the analysis. Some of the clinical information, e.g., race/ethnicity of the patients, was not recorded for 47% of the patients, and some CNV may be associated more with different nationalities/backgrounds (39). The retrospective analysis only allowed the ability to compare genetic alterations with the presence of metastatic disease. The inability to have a prospective component after genetic analysis limits the application of the findings and prevents the determination of factors associated with development of distant metastases.
Although other studies have shown somatic changes and CNV that characterize the genomes of multiple cancer types, no previous study has associated the CNV of TNFRSF10C at the primary site with distant metastatic disease. The association could influence treatment strategies, as well as potentially serve as a marker for targeted molecular therapy.
Conclusions
In conclusion, our cohort displayed a statistically significant association between TNFRSF10C CNV and distant metastatic disease in CRC and rectal adenocarcinoma. With further validation in longitudinal studies, TNFRSF10C CNV may be used clinically to support optimal systemic treatment strategies versus more aggressive local therapies in patients with CRC, including radiation therapy for rectal adenocarcinoma.
Acknowledgements
Funding: This work was supported in part by the Biostatistics and Bioinformatics Shared resource of Winship Cancer Institute of Emory University and Nation Institutes of Health/National Cancer Institute under award number P30CA138292.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Disclaimer: The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin 2014;64:104-17. [Crossref] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330-7. [Crossref] [PubMed]
- Fodde R, Smits R, Clevers H. APC, signal transduction and genetic instability in colorectal cancer. Nat Rev Cancer 2001;1:55-67. [Crossref] [PubMed]
- de la Chapelle A. Genetic predisposition to colorectal cancer. Nat Rev Cancer 2004;4:769-80. [Crossref] [PubMed]
- Improved survival with preoperative radiotherapy in resectable rectal cancer. Swedish Rectal Cancer Trial. N Engl J Med 1997;336:980-7. [Crossref] [PubMed]
- Tonini GP, Boni L, Pession A, et al. MYCN oncogene amplification in neuroblastoma is associated with worse prognosis, except in stage 4s: the Italian experience with 295 children. J Clin Oncol 1997;15:85-93. [PubMed]
- Vakiani E, Janakiraman M, Shen R, et al. Comparative genomic analysis of primary versus metastatic colorectal carcinomas. J Clin Oncol 2012;30:2956-62. [Crossref] [PubMed]
- Lee H, Flaherty P, Ji HP. Systematic genomic identification of colorectal cancer genes delineating advanced from early clinical stage and metastasis. BMC Med Genomics 2013;6:54. [Crossref] [PubMed]
- Xie T, Cho YB, Wang K, et al. Patterns of somatic alterations between matched primary and metastatic colorectal tumors characterized by whole-genome sequencing. Genomics 2014;104:234-41. [Crossref] [PubMed]
- Lee SY, Haq F, Kim D, et al. Comparative genomic analysis of primary and synchronous metastatic colorectal cancers. PLoS One 2014;9:e90459. [Crossref] [PubMed]
- Mekenkamp LJ, Haan JC, Israeli D, et al. Chromosomal copy number aberrations in colorectal metastases resemble their primary counterparts and differences are typically non-recurrent. PLoS One 2014;9:e86833. [Crossref] [PubMed]
- González-González M, Fontanillo C, Abad MM, et al. Identification of a characteristic copy number alteration profile by high-resolution single nucleotide polymorphism arrays associated with metastatic sporadic colorectal cancer. Cancer 2014;120:1948-59. [Crossref] [PubMed]
- Macartney-Coxson DP, Hood KA, Shi HJ, et al. Metastatic susceptibility locus, an 8p hot-spot for tumour progression disrupted in colorectal liver metastases: 13 candidate genes examined at the DNA, mRNA and protein level. BMC Cancer 2008;8:187. [Crossref] [PubMed]
- Cheng Y, Kim JW, Liu W, et al. Genetic and epigenetic inactivation of TNFRSF10C in human prostate cancer. Prostate 2009;69:327-35. [Crossref] [PubMed]
- Shivapurkar N, Toyooka S, Toyooka KO, et al. Aberrant methylation of trail decoy receptor genes is frequent in multiple tumor types. Int J Cancer 2004;109:786-92. [Crossref] [PubMed]
- Chughtai SA, Crundwell MC, Cruickshank NR, et al. Two novel regions of interstitial deletion on chromosome 8p in colorectal cancer. Oncogene 1999;18:657-65. [Crossref] [PubMed]
- Falschlehner C, Emmerich CH, Gerlach B, et al. TRAIL signalling: decisions between life and death. Int J Biochem Cell Biol 2007;39:1462-75. [Crossref] [PubMed]
- Oikonomou E, Kosmidou V, Katseli A, et al. TRAIL receptor upregulation and the implication of KRAS/BRAF mutations in human colon cancer tumors. Int J Cancer 2009;125:2127-35. [Crossref] [PubMed]
- Granci V, Bibeau F, Kramar A, et al. Prognostic significance of TRAIL-R1 and TRAIL-R3 expression in metastatic colorectal carcinomas. Eur J Cancer 2008;44:2312-8. [Crossref] [PubMed]
- Takayama T, Miyanishi K, Hayashi T, et al. Colorectal cancer: genetics of development and metastasis. J Gastroenterol 2006;41:185-92. [Crossref] [PubMed]
- Sträter J, Hinz U, Walczak H, et al. Expression of TRAIL and TRAIL receptors in colon carcinoma: TRAIL-R1 is an independent prognostic parameter. Clin Cancer Res 2002;8:3734-40. [PubMed]
- Ganten TM, Sykora J, Koschny R, et al. Prognostic significance of tumour necrosis factor-related apoptosis-inducing ligand (TRAIL) receptor expression in patients with breast cancer. J Mol Med (Berl) 2009;87:995-1007. [Crossref] [PubMed]
- van Geelen CM, Westra JL, de Vries EG, et al. Prognostic significance of tumor necrosis factor-related apoptosis-inducing ligand and its receptors in adjuvantly treated stage III colon cancer patients. J Clin Oncol 2006;24:4998-5004. [Crossref] [PubMed]
- Kuijlen JM, Mooij JJ, Platteel I, et al. TRAIL-receptor expression is an independent prognostic factor for survival in patients with a primary glioblastoma multiforme. J Neurooncol 2006;78:161-71. [Crossref] [PubMed]
- Johnstone RW, Frew AJ, Smyth MJ. The TRAIL apoptotic pathway in cancer onset, progression and therapy. Nat Rev Cancer 2008;8:782-98. [Crossref] [PubMed]
- Bellail AC, Qi L, Mulligan P, et al. TRAIL agonists on clinical trials for cancer therapy: the promises and the challenges. Rev Recent Clin Trials 2009;4:34-41. [Crossref] [PubMed]
- Wainberg ZA, Messersmith WA, Peddi PF, et al. A phase 1B study of dulanermin in combination with modified FOLFOX6 plus bevacizumab in patients with metastatic colorectal cancer. Clin Colorectal Cancer 2013;12:248-54. [Crossref] [PubMed]
- Trarbach T, Moehler M, Heinemann V, et al. Phase II trial of mapatumumab, a fully human agonistic monoclonal antibody that targets and activates the tumour necrosis factor apoptosis-inducing ligand receptor-1 (TRAIL-R1), in patients with refractory colorectal cancer. Br J Cancer 2010;102:506-12. [Crossref] [PubMed]
- Wiezorek J, Holland P, Graves J. Death receptor agonists as a targeted therapy for cancer. Clin Cancer Res 2010;16:1701-8. [Crossref] [PubMed]
- Alliance for Clinical Trials in Oncology, National Cancer Institute (NCI). Chemotherapy Alone or Chemotherapy Plus Radiation Therapy in Treating Patients With Locally Advanced Rectal Cancer Undergoing Surgery. 2015. Available online: https://clinicaltrials.gov/ct2/show/NCT01515787
- Malesci A, Laghi L, Bianchi P, et al. Reduced likelihood of metastases in patients with microsatellite-unstable colorectal cancer. Clin Cancer Res 2007;13:3831-9. [Crossref] [PubMed]
- Missiaglia E, Jacobs B, D'Ario G, et al. Distal and proximal colon cancers differ in terms of molecular, pathological, and clinical features. Ann Oncol 2014;25:1995-2001. [Crossref] [PubMed]
- Sugai T, Habano W, Jiao YF, et al. Analysis of molecular alterations in left- and right-sided colorectal carcinomas reveals distinct pathways of carcinogenesis: proposal for new molecular profile of colorectal carcinomas. J Mol Diagn 2006;8:193-201. [Crossref] [PubMed]
- Frattini M, Balestra D, Suardi S, et al. Different genetic features associated with colon and rectal carcinogenesis. Clin Cancer Res 2004;10:4015-21. [Crossref] [PubMed]
- Kapiteijn E, Liefers GJ, Los LC, et al. Mechanisms of oncogenesis in colon versus rectal cancer. J Pathol 2001;195:171-8. [Crossref] [PubMed]
- Minsky BD. Unique considerations in the patient with rectal cancer. Semin Oncol 2011;38:542-51. [Crossref] [PubMed]
- Sarli L, Bader G, Iusco D, et al. Number of lymph nodes examined and prognosis of TNM stage II colorectal cancer. Eur J Cancer 2005;41:272-9. [Crossref] [PubMed]
- Xingmao Z, Hongying W, Zhixiang Z, et al. Analysis on the correlation between number of lymph nodes examined and prognosis in patients with stage II colorectal cancer. Med Oncol 2013;30:371. [Crossref] [PubMed]
- Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature 2006;444:444-54. [Crossref] [PubMed]


